CXCR4+ mammary gland macrophageal niche promotes tumor initiating cell activity and immune suppression during tumorigenesis
- PMID: 40413176
- PMCID: PMC12103607
- DOI: 10.1038/s41467-025-59972-z
CXCR4+ mammary gland macrophageal niche promotes tumor initiating cell activity and immune suppression during tumorigenesis
Abstract
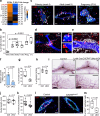
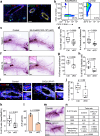
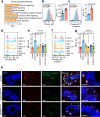
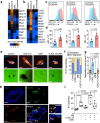
Tumor-initiating cells (TICs) share features and regulatory pathways with normal stem cells, yet how the stem cell niche contributes to tumorigenesis remains unclear. Here, we identify CXCR4+ macrophages as a niche population enriched in normal mammary ducts, where they promote the regenerative activity of basal cells in response to luminal cell-derived CXCL12. CXCL12 triggers AKT-mediated stabilization of β-catenin, which induces Wnt ligands and pro-migratory genes, enabling intraductal macrophage infiltration and supporting regenerative activity of basal cells. Notably, these same CXCR4+ niche macrophages regulate the tumor-initiating activity of various breast cancer subtypes by enhancing TIC survival and tumor-forming capacity, while promoting early immune evasion through regulatory T cell induction. Furthermore, a CXCR4+ niche macrophage gene signature correlates with poor prognosis in human breast cancer. These findings highlight the pivotal role of the CXCL12-CXCR4 axis in orchestrating interactions between niche macrophages, mammary epithelial cells, and immune cells, thereby establishing a supportive niche for both normal tissue regeneration and mammary tumor initiation.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: Y.K. is a co-founder and chair of the Scientific Advisory Board of Firebrand Therapeutics, Inc. and Kayothera, Inc. The remaining authors declare no competing interests.
Figures



References
-
- Clarke, M. F. et al. Cancer stem cells-perspectives on current status and future directions: AACR Workshop on cancer stem cells. Cancer Res.66, 9339–9344 (2006). - PubMed
-
- Kreso, A. & Dick, J. E. Evolution of the cancer stem cell model. Cell Stem Cell14, 275–291 (2014). - PubMed
-
- Nguyen, L. V., Vanner, R., Dirks, P. & Eaves, C. J. Cancer stem cells: an evolving concept. Nat. Rev. Cancer12, 133–143 (2012). - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

