Microevolution and genomic epidemiology of the diphtheria-causing zoonotic pathogen Corynebacterium ulcerans
- PMID: 40413184
- PMCID: PMC12103533
- DOI: 10.1038/s41467-025-60065-0
Microevolution and genomic epidemiology of the diphtheria-causing zoonotic pathogen Corynebacterium ulcerans
Abstract
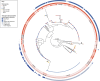
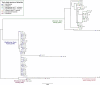
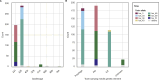
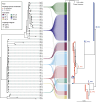
Corynebacterium ulcerans is an emerging zoonotic pathogen which causes diphtheria-like infections. Although C. ulcerans is found in multiple domestic and wild animal species, most human cases are linked with pets. Our ability to decipher cross-host species transmission dynamics and to understand the emergence of clinically relevant clones (e.g., diphtheria toxin-positive) is currently hampered by a limited knowledge of C. ulcerans strain diversity and genome evolution. Here, we explore the genomic population structure and evolution of C. ulcerans with 582 isolates from diverse hosts and geographical locations. A newly developed core genome genotyping scheme captures the population structure of C. ulcerans both at deep and shallow phylogenetic levels, uncovering its main sublineages and offering high strain subtyping resolution for epidemiological surveillance. Additionally, we reveal the diversity and distribution of the diphtheria toxin gene (tox), and those of its associated mobile elements. Considering the entire Corynebacterium diphtheriae Species Complex, we find four diphtheria toxin families, five tox-prophage families, and a novel tox-carrying genetic element. We show that some toxin families are shared across Corynebacterium species, revealing tox-prophage cross-species transfer. Our work enhances knowledge on the ecology and evolution of C. ulcerans and provides a genomic framework for tracking the dissemination of emerging sublineages.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures


References
-
- Moore, L. S. P. et al. Corynebacterium ulcerans cutaneous diphtheria. Lancet Infect. Dis.15, 1100–1107 (2015). - PubMed
-
- Dias, A. A. et al. Corynebacteriumulcerans diphtheria: an emerging zoonosis in Brazil and worldwide. Rev. Saúde Pública45, 1176–1191 (2011). - PubMed
-
- Hacker, E., Antunes, C. A., Mattos-Guaraldi, A. L., Burkovski, A. & Tauch, A. Corynebacterium ulcerans, an emerging human pathogen. Future Microbiol.11, 1191–1208 (2016). - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

