Analysis of more than 400,000 women provides case-control evidence for BRCA1 and BRCA2 variant classification
- PMID: 40413188
- PMCID: PMC12103537
- DOI: 10.1038/s41467-025-59979-6
Analysis of more than 400,000 women provides case-control evidence for BRCA1 and BRCA2 variant classification
Abstract
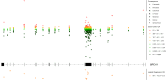
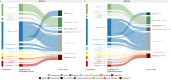
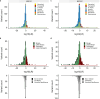
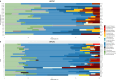
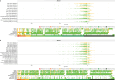
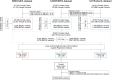
Clinical genetic testing identifies variants causal for hereditary cancer, information that is used for risk assessment and clinical management. Unfortunately, some variants identified are of uncertain clinical significance (VUS), complicating patient management. Case-control data is one evidence type used to classify VUS. As an initiative of the Evidence-based Network for the Interpretation of Germline Mutant Alleles (ENIGMA) Analytical Working Group we analyze germline sequencing data of BRCA1 and BRCA2 from 96,691 female breast cancer cases and 302,116 controls from three studies: the BRIDGES study of the Breast Cancer Association Consortium, the Cancer Risk Estimates Related to Susceptibility consortium, and the UK Biobank. We observe 11,207 BRCA1 and BRCA2 variants, with 6909 being coding, covering 23.4% of BRCA1 and BRCA2 VUS in ClinVar and 19.2% of ClinVar curated (likely) benign or pathogenic variants. Case-control likelihood ratio (ccLR) evidence is highly consistent with ClinVar assertions for (likely) benign or pathogenic variants; exhibiting 99.1% sensitivity and 95.3% specificity for BRCA1 and 93.3% sensitivity and 86.6% specificity for BRCA2. This approach provides case-control evidence for 787 unclassified variants; these include 579 with strong or moderate benign evidence and 10 with strong pathogenic evidence for which ccLR evidence is sufficient to alter clinical classification.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The following authors declare conflicts not directly relevant to this work as stated below. VK-MT and GDE have received consultancies from AstraZeneca and Everything Genetic Ltd. ACA and DFE are named creators of the BOADICEA model, which has been licensed by Cambridge Enterprise (University of Cambridge). SJH has received speaker fees from AstraZeneca and Pfizer Ltd. SY has received research funding from AstraZeneca and Repare Therapeutics and participates in the advisory board for AstraZeneca. JWW has received consulting fees from Natera, MyOme, Cancer IQ, equity from Natera, and speaker fees from AstraZeneca. All the other authors declare no competing interests.
Figures


Update of
-
Analysis of more than 400,000 women provides case-control evidence for BRCA1 and BRCA2 variant classification.medRxiv [Preprint]. 2024 Sep 4:2024.09.04.24313051. doi: 10.1101/2024.09.04.24313051. medRxiv. 2024. Update in: Nat Commun. 2025 May 25;16(1):4852. doi: 10.1038/s41467-025-59979-6. PMID: 39281752 Free PMC article. Updated. Preprint.
References
MeSH terms
Substances
Grants and funding
- HHSN261201800009C/CA/NCI NIH HHS/United States
- UM1 CA176726/CA/NCI NIH HHS/United States
- R37 CA070867/CA/NCI NIH HHS/United States
- P30 CA014520/CA/NCI NIH HHS/United States
- HHSN261201800015I/CA/NCI NIH HHS/United States
- HHSN261201000091C/CA/NCI NIH HHS/United States
- HHSN261201800016C/CA/NCI NIH HHS/United States
- 75N92021D00001/HL/NHLBI NIH HHS/United States
- U01 CA167551/CA/NCI NIH HHS/United States
- R01 CA064277/CA/NCI NIH HHS/United States
- P50 CA116201/CA/NCI NIH HHS/United States
- R35 CA253187/CA/NCI NIH HHS/United States
- HHSN261201800032I/CA/NCI NIH HHS/United States
- R01 CA264971/CA/NCI NIH HHS/United States
- U01 CA199277/CA/NCI NIH HHS/United States
- R01 CA225662/CA/NCI NIH HHS/United States
- R01 CA163353/CA/NCI NIH HHS/United States
- UM1 CA164917/CA/NCI NIH HHS/United States
- HHSN261201800032C/CA/NCI NIH HHS/United States
- 75N92021D00002/HL/NHLBI NIH HHS/United States
- NU58DP006344/DP/NCCDPHP CDC HHS/United States
- 75N92021D00005/WH/WHI NIH HHS/United States
- HHSN261201800011C/CA/NCI NIH HHS/United States
- P01 CA087969/CA/NCI NIH HHS/United States
- U01 CA164974/CA/NCI NIH HHS/United States
- R01 CA098663/CA/NCI NIH HHS/United States
- U01 CA242954/CA/NCI NIH HHS/United States
- R01 CA148667/CA/NCI NIH HHS/United States
- HHSN261201800021C/CA/NCI NIH HHS/United States
- HHSN261201800016I/CA/NCI NIH HHS/United States
- 75N92021D00003/WH/WHI NIH HHS/United States
- UM1 CA186107/CA/NCI NIH HHS/United States
- P30 CA023100/CA/NCI NIH HHS/United States
- R01 CA077398/CA/NCI NIH HHS/United States
- P30 CA068485/CA/NCI NIH HHS/United States
- U01 CA164920/CA/NCI NIH HHS/United States
- U19 CA148065/CA/NCI NIH HHS/United States
- P30 CA033572/CA/NCI NIH HHS/United States
- WT_/Wellcome Trust/United Kingdom
- HHSN261201800015C/CA/NCI NIH HHS/United States
- HHSN261201800001C/CA/NCI NIH HHS/United States
- HHSN261201800031C/CA/NCI NIH HHS/United States
- U01 CA164973/CA/NCI NIH HHS/United States
- 75N92021D00004/WH/WHI NIH HHS/United States
- U24 CA258058/CA/NCI NIH HHS/United States
- R01 CA185623/CA/NCI NIH HHS/United States
- P30 CA042014/CA/NCI NIH HHS/United States
- HHSN261201800009I/CA/NCI NIH HHS/United States
- R01 CA100598/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

