Gene2role: a role-based gene embedding method for comparative analysis of signed gene regulatory networks
- PMID: 40413377
- PMCID: PMC12103023
- DOI: 10.1186/s12859-025-06128-x
Gene2role: a role-based gene embedding method for comparative analysis of signed gene regulatory networks
Abstract
Background: Understanding the dynamics of gene regulatory networks (GRNs) across various cellular states is crucial for deciphering the underlying mechanisms governing cell behavior and functionality. However, current comparative analytical methods, which often focus on simple topological information such as the degree of genes, are limited in their ability to fully capture the similarities and differences among the complex GRNs.
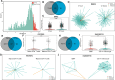
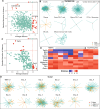
Results: We present Gene2role, a gene embedding approach that leverages multi-hop topological information from genes within signed GRNs. Initially, we demonstrated the effectiveness of Gene2role in capturing the intricate topological nuances of genes using GRNs inferred from four distinct data sources. Then, applying Gene2role to integrated GRNs allowed us to identify genes with significant topological changes across cell types or states, offering a fresh perspective beyond traditional differential gene expression analyses. Additionally, we quantified the stability of gene modules between two cellular states by measuring the changes in the gene embeddings within these modules.
Conclusions: Our method augments the existing toolkit for probing the dynamic regulatory landscape, thereby opening new avenues for understanding gene behavior and interaction patterns across cellular transitions.
Keywords: Gene module analysis; Gene regulatory network analysis; Graph representation learning.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Ethics approval and consent to participate: Not applicable. Consent for publication: Not applicable. Competing interests: The authors declare no competing interests.
Figures



Similar articles
-
MICRAT: a novel algorithm for inferring gene regulatory networks using time series gene expression data.BMC Syst Biol. 2018 Dec 14;12(Suppl 7):115. doi: 10.1186/s12918-018-0635-1. BMC Syst Biol. 2018. PMID: 30547796 Free PMC article.
-
scMGATGRN: a multiview graph attention network-based method for inferring gene regulatory networks from single-cell transcriptomic data.Brief Bioinform. 2024 Sep 23;25(6):bbae526. doi: 10.1093/bib/bbae526. Brief Bioinform. 2024. PMID: 39417321 Free PMC article.
-
AttentionGRN: a functional and directed graph transformer for gene regulatory network reconstruction from scRNA-seq data.Brief Bioinform. 2025 Mar 4;26(2):bbaf118. doi: 10.1093/bib/bbaf118. Brief Bioinform. 2025. PMID: 40116659 Free PMC article.
-
Learning Differential Module Networks Across Multiple Experimental Conditions.Methods Mol Biol. 2019;1883:303-321. doi: 10.1007/978-1-4939-8882-2_13. Methods Mol Biol. 2019. PMID: 30547406 Review.
-
Computational methods for discovering gene networks from expression data.Brief Bioinform. 2009 Jul;10(4):408-23. doi: 10.1093/bib/bbp028. Brief Bioinform. 2009. PMID: 19505889 Review.
References
-
- Akers K, Murali TM. Gene regulatory network inference in single-cell biology. Curr Opin Syst Biol. 2021;26:87–97.
-
- Badia-i-Mompel P, Wessels L, Müller-Dott S, Trimbour R, Ramirez Flores RO, Argelaguet R, et al. Gene regulatory network inference in the era of single-cell multi-omics. Nat Rev Genet. 2023;24(11):739–54. - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

