A novel KRAS exon 2 drop-off digital PCR assay for mutation detection in cell-free DNA of cancer patients
- PMID: 40413426
- PMCID: PMC12103757
- DOI: 10.1186/s13000-025-01637-y
A novel KRAS exon 2 drop-off digital PCR assay for mutation detection in cell-free DNA of cancer patients
Abstract
Background: KRAS exon 2 mutations are highly prevalent in human malignancies, making them attractive targets for detection and monitoring in cell-free DNA (cfDNA) of cancer patients. Drop-off assays designed for digital polymerase chain reaction (ddPCR drop-off) span entire mutational hotspots and detect any mutated allele within the covered region, overcoming a major limitation of mutation-specific ddPCR assays. We therefore set out to develop a novel KRAS codon 12/13 ddPCR drop-off assay for the robust, highly sensitive and specific detection of KRAS exon 2 hotspot mutations in cfDNA.
Methods: We designed, optimized and extensively validated a KRAS codon 12/13 ddPCR drop-off assay. We compared assay performance to a commercially available KRAS multiplex assay. For clinical validation, we analyzed plasma samples collected from patients with KRAS-mutated gastrointestinal malignancies.
Results: Limit of detection of the newly established ddPCR drop-off assay was 0.57 copies/µL, limit of blank was 0.13 copies/µ. The inter-assay precision (r2) was 0.9096. Our newly developed KRAS ddPCR drop-off assay accurately identified single nucleotide variants in 35/36 (97.2%) of circulating tumor DNA-positive samples from the patient validation cohort. Assay cross-validation showed that the newly established KRAS codon 12/13 ddPCR drop-off assay outperformed a commercially available KRAS multiplex ddPCR assay in terms of specificity. Moreover, the newly developed assay proved to be suitable for multiplexing with mutation-specific probes.
Conclusion: We developed and clinically validated a highly accurate ddPCR drop-off assay for KRAS exon 2 hot-spot detection in cfDNA with broad applicability for clinic and research.
Keywords: KRAS; Cell-free DNA (cfDNA); Circulating-tumor DNA (ctDNA); Drop-off assay; Droplet digital polymerase chain reaction (ddPCR); Liquid biopsy; Precision medicine.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Ethics approval and consent to participate: The study was approved by the local institutional review boards (IRB) in Freiburg (EK-Freiburg project number 46/18) and Zurich (BASEC-Nr. 2020-01 104). All patients gave written informed consent for collection and analysis of blood samples. No identifiable images or data are included in this manuscript. All study procedures were conducted in accordance with the Good Clinical Practice guidelines and the Declaration of Helsinki. Consent for publication: Not applicable. Competing interests: The authors declare no competing interests.
Figures
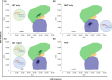
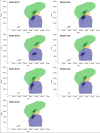
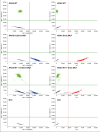


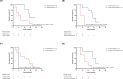
Similar articles
-
Development and Clinical Validation of Discriminatory Multitarget Digital Droplet PCR Assays for the Detection of Hot Spot KRAS and NRAS Mutations in Cell-Free DNA.J Mol Diagn. 2020 Jul;22(7):943-956. doi: 10.1016/j.jmoldx.2020.04.206. Epub 2020 May 4. J Mol Diagn. 2020. PMID: 32376474
-
Multiplex KRASG12/G13 mutation testing of unamplified cell-free DNA from the plasma of patients with advanced cancers using droplet digital polymerase chain reaction.Ann Oncol. 2017 Mar 1;28(3):642-650. doi: 10.1093/annonc/mdw670. Ann Oncol. 2017. PMID: 27993791 Free PMC article.
-
Development of Highly Sensitive Digital Droplet PCR for Detection of cKIT Mutations in Circulating Free DNA That Mediate Resistance to TKI Treatment for Gastrointestinal Stromal Tumor (GIST).Int J Mol Sci. 2023 Mar 12;24(6):5411. doi: 10.3390/ijms24065411. Int J Mol Sci. 2023. PMID: 36982486 Free PMC article.
-
Performance of four platforms for KRAS mutation detection in plasma cell-free DNA: ddPCR, Idylla, COBAS z480 and BEAMing.Sci Rep. 2020 May 15;10(1):8122. doi: 10.1038/s41598-020-64822-7. Sci Rep. 2020. PMID: 32415199 Free PMC article.
-
Cell-free DNA for the detection of pancreatic, liver and upper gastrointestinal cancers: has progress been made?Future Oncol. 2013 Dec;9(12):1861-9. doi: 10.2217/fon.13.152. Future Oncol. 2013. PMID: 24295416 Review.
References
-
- Alix-Panabières C, Pantel K. Clinical applications of Circulating tumor cells and Circulating tumor DNA as liquid biopsy. Cancer Discov. 2016;6:479–91. 10.1158/2159-8290.Cd-15-1483. - PubMed
-
- Wan JCM, et al. Liquid biopsies come of age: towards implementation of Circulating tumour DNA. Nat Rev Cancer. 2017;17:223–38. 10.1038/nrc.2017.7. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

