Translating Muscle RNAseq Into the Clinic for the Diagnosis of Muscle Diseases
- PMID: 40413734
- PMCID: PMC12257123
- DOI: 10.1002/acn3.70078
Translating Muscle RNAseq Into the Clinic for the Diagnosis of Muscle Diseases
Abstract
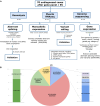
Objective: Approximately half of patients with hereditary myopathies remain without a definitive genetic diagnosis after DNA next-generation sequencing (NGS). Here, we implemented transcriptome analysis of muscle biopsies as a complementary diagnostic tool for patients with muscle disease but no definitive genetic diagnosis after exome sequencing.
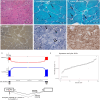
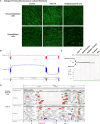
Methods: In total, 70 undiagnosed cases with suspected genetic muscular dystrophies or congenital myopathies were included in the study. Muscle RNAseq comprised the analysis of aberrant splicing, aberrant expression, and monoallelic expression. In addition, existing NGS data or variant calling from RNAseq were reanalyzed, and genome sequencing was performed in selected cases. Four aberrant splicing open-source tools were compared and assessed.
Results: RNAseq established a diagnosis in 10/70 patients (14.3%) by identifying aberrant transcripts produced by single nucleotide variants (7/10) or copy number variants (3/10). Reanalysis of NGS data allowed the diagnosis in 9/70 individuals (12.9%). Based on this cohort, FRASER was the tool that reported more splicing outlier events per sample while showing the highest accuracy (81.26%).
Conclusions: We demonstrate the utility of RNAseq in identifying causative variants in muscle diseases. Evaluation of four aberrant splicing tools allowed efficient identification of most pathogenic splicing events, obtaining a manageable number of candidate events for manual inspection, demonstrating feasibility for translation into a clinical setting. We also show how the integration of omic technologies reduces the turnaround time to identify causative variants.
Keywords: RNA sequencing; alternative splicing; congenital myopathy; genetic diagnosis; muscular dystrophy; neuromuscular diseases; transcriptomics.
© 2025 The Author(s). Annals of Clinical and Translational Neurology published by Wiley Periodicals LLC on behalf of American Neurological Association.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures

References
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
