PRMT1 inhibitor MS023 suppresses RNA splicing to sensitize small cell lung cancer to DNA damaging agents
- PMID: 40413955
- PMCID: PMC12152348
- DOI: 10.1016/j.neo.2025.101176
PRMT1 inhibitor MS023 suppresses RNA splicing to sensitize small cell lung cancer to DNA damaging agents
Abstract
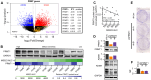
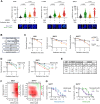
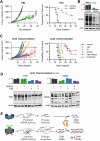
Small cell lung cancer (SCLC) is a highly aggressive form of cancer, commonly treated with DNA-damaging therapies such as chemotherapy and radiotherapy. Unfortunately, relapse occurs early and frequently, suggesting that epigenetic mechanisms may play a role in this aggressive behavior. Targeting these mechanisms during initial treatment could potentially enhance anti-cancer effects. This study investigated the combination of DNA-damaging treatments with a panel of Epigenetic Chemical Probes (EpiProbes). Among these, MS023, a PRMT inhibitor, showed the greatest synergy with cisplatin and etoposide across various SCLC cell lines. The cytotoxicity of MS023 was correlated with PRMT1 gene expression and protein levels. BioID analysis revealed that many PRMT1 interactors are involved in mRNA splicing. Mechanistic validation demonstrated that MS023 impaired RNA splicing, increased DNA:RNA hybrids, and caused DNA double-strand breaks (DSBs). When combined with ionizing radiation (IR), MS023 significantly increased DSBs, as indicated by γH2AX foci. Additionally, MS023 enhanced the effects of IR and the PARP inhibitor talazoparib, both in vitro and in vivo. Therefore, targeting PRMT1 in combination with DNA-damaging therapies presents a promising strategy to improve treatment outcomes for SCLC.
Keywords: Chemotherapy; Drug synergy; Epigenetics; PARP inhibitor; Radiation therapy; Small cell lung cancer, Poly(ADP-Ribose) polymerase.
Copyright © 2025. Published by Elsevier Inc.
Conflict of interest statement
Declaration of competing interest The authors declare the following financial interests/personal relationships which may be considered as potential competing interests: B.H. Lok reports grants from Pfizer and grants, personal fees, and nonfinancial support from AstraZeneca, and personal fees from Daiichi-Sankyo outside the submitted work. No other co-authors have any conflicts of interests to declare.
Figures


References
-
- Bray F., Ferlay J., Laversanne M., Brewster D.H., Gombe Mbalawa C., Kohler B., Piñeros M., Steliarova-Foucher E., Swaminathan R., Antoni S., et al. Cancer I ncidence in F ive C ontinents: Inclusion criteria, highlights from Volume X and the global status of cancer registration. Int. J. Cancer. 2015;137:2060–2071. doi: 10.1002/ijc.29670. - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

