Conjugated bile acids promote metabolic dysfunction-associated steatotic liver disease through inducing nuclear translocation of sphingosine-1-phosphate receptor 2 to disrupt peroxisome proliferator-activated receptor alpha
- PMID: 40414852
- PMCID: PMC12103792
- DOI: 10.1186/s12964-025-02249-1
Conjugated bile acids promote metabolic dysfunction-associated steatotic liver disease through inducing nuclear translocation of sphingosine-1-phosphate receptor 2 to disrupt peroxisome proliferator-activated receptor alpha
Abstract
Background: Conjugated bile acids (CBAs) induced metabolic dysfunction-associated steatotic liver disease (MASLD) through activating sphingosine-1-phosphate receptor 2 (S1PR2). However, the precise mechanisms have not been fully understood.
Methods: We established a link between CBAs and MASLD in IBD patients with high SphK2 and Villin-SphK2TG mice. Villin-SphK2TG mice were fed high-fat diet (HFD) for inducing MASLD. Gut microbiota composition was analyzed by 16 S rDNA Amplicon sequencing assay. We performed the UPLC/TQMS based targeted metabolomics assay to analyze the compositions of BAs in liver, serum and feces. In vitro assays, hepatocytes transfected with N-terminal truncated-S1PR2 analyzed the dynamics of S1PR2 stimulated by CBAs. PPARα function was assayed by analyzing the DNA-protein interactions by using Electrophoretic mobility shift assay (EMSA).
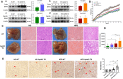
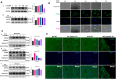
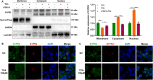
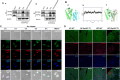
Results: The IBD patients with high colonocyte SphK2 conferred the development of MASLD. Feeding high-fat diet, Villin-SphK2TG mice developed MASLD more severely than WT mice. Analysis of gut microbiota showed that colonic SphK2 shaped microbiota by reducing the BSH-producing bacteria, thus leading to the accumulation of CBAs in liver via the gut-liver axis. CBAs induced nuclear translocation of S1PR2 through cleavage the N-terminal sequences of Ala-Ser-Ala-Phe-Iso in hepatocytes. Cleaved S1PR2 (S1PR2') was thus translocated into the nucleus to bind with PPARα, thereby interdicting the function of PPARα in regulating the genes involved in lipid catabolism. S1PR2 antagonist JTE-013 blocked the CBAs-induced nuclear translocation of S1PR2 and S1PR2 is thus identified as a potential therapeutic target for MASLD treatment.
Conclusion: CBAs promoted MASLD through inducing S1PR2 translocation into the nucleus, where it bound PPARα to interdict the function of PPARα in regulating genes involved in lipid catabolism.
Keywords: Conjugated bile acid (CBAs); MASLD; Nuclear translocation of S1PR2; PPARα; S1PR2.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Ethical approval: Ethics Committee of Binzhou Medical College Hospital approved the use of human hepatocellular carcinoma samples (No.2020-012). Animal Welfare Committee of Capital Medical University approved the protocols of animal studies (AEEI-2019–043). Consent for publication: Yes. Competing interests: The authors declare no competing interests.
Figures


References
-
- Chavez-Talavera O, Tailleux A, Lefebvre P, Staels B. Bile acid control of metabolism and inflammation in obesity, type 2 diabetes, dyslipidemia, and nonalcoholic fatty liver disease. Gastroenterology. 2017;152:1679–94. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

