CD24 Regulates the Formation of Ectosomes in B Lymphocytes
- PMID: 40415253
- PMCID: PMC12104215
- DOI: 10.1002/jev2.70093
CD24 Regulates the Formation of Ectosomes in B Lymphocytes
Abstract
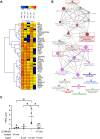
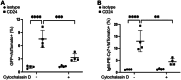
CD24 is a glycophosphatidylinositol-linked protein that regulates B cell development. We previously reported that stimulation of CD24 on donor B cells promotes the transfer of functional receptors to recipient B cells via extracellular vesicles (EVs). However, the mechanisms regulating CD24-mediated formation of bioactive EVs are unknown. Using bioinformatics, we found a connection between CD24, and PI3K/AKT, tran and mTOR. To determine if these pathways regulate EV release, we used flow cytometry to follow the transfer of EVs carrying lipid-associated GFP and surface IgM from donor to recipient B cells. Using chemical and genetic inhibition, we found that a PI3K/mTORC2/ROCK/actin pathway regulates bioactive EV formation via activation of acid sphingomyelinase (aSMase) upstream of PI3K. Using single EV analysis, we found that CD24 regulates the formation of the subset of bioactive EVs that are taken up by recipient cells and not total EVs. Interestingly, we also found that ROCK and aSMase modulate ectosome but not exosome formation, when CD24 is stimulated. Lastly, through live cell imaging, we found that PI3K and ROCK are required for inducing membrane dynamics associated with EV formation. These data suggest that this pathway regulates bioactive EV release that, in turn, could regulate B cell development.
Keywords: B lymphocyte; CD24; acid sphingomyelinase (aSMase); ceramide; ectosome; extracellular vesicle.
© 2025 The Author(s). Journal of Extracellular Vesicles published by Wiley Periodicals LLC on behalf of International Society for Extracellular Vesicles.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures






References
-
- Brás, I. C. , Khani M. H., Riedel D., et al. 2022. “Ectosomes and Exosomes Modulate Neuronal Spontaneous Activity.” Journal of Proteomics 269: 104721. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

