Probiotic efficacy and mechanism of a pigeon derived Ligilactobacillus salivarius strain in promoting growth and intestinal development of pigeons
- PMID: 40415945
- PMCID: PMC12098536
- DOI: 10.3389/fmicb.2025.1584380
Probiotic efficacy and mechanism of a pigeon derived Ligilactobacillus salivarius strain in promoting growth and intestinal development of pigeons
Abstract
Background: With the gradual rise of antibiotic-free farming practices, the exploration of novel, green, and low-pollution alternatives to antibiotics has become one of the key research focus in the field of agricultural science. In the development of antibiotic alternatives, probiotics, particularly host-associated probiotics, have been found to play a significant role in enhancing the production performance of livestock and poultry. However, research on and application of probiotics specifically for meat pigeons remain relatively underdeveloped.
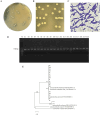
Objective: To assess and investigate the probiotic efficacy and mechanisms during homologous lactic acid bacteria (LAB) transplant to host-pigeons, LAB strains with good probiotic properties were isolated from the intestinal contents of 28-day-old Mimas pigeons. And then measured the production indexes, intestinal flora, and intestinal transcriptomics of the hosts after instillation of LAB strains.
Methods: A total of 360 at 1-day-old pigeons were randomly divided into four groups and gavaged 0.4 mL Ligilactobacillus salivarius S10 with concentration of 0, 108, 109, and 1010 CFU/mL, designated as the control group (CG), the low concentration group (LG), the medium concentration group (MG), and the high concentration group (HG), respectively.
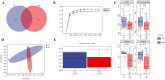
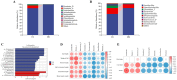
Results: The findings revealed that an optimal concentration of 109 CFU/mL L. salivarius S10, a dominant strain isolated and screened, enhanced the growth performance and intestinal development of young pigeons. 16S rRNA gene sequencing analysis demonstrated a significant increase in the abundance of Lactobacillus, Pantoea_A and Enterococcus_H and a significant reduction in the abundance of Clostridium_T in the pigeon ileum (p < 0.05) under selected concentration treatment. Transcriptomic profiling of the ileum revealed 1828 differentially expressed genes (DEGs) between CG and MG. Notably, DEGs involved in the MAPK signaling pathway, such as RAF1, PDGFRB, and ELK4, were significantly correlated with differential ileal bacteria, suggesting that modulation of intestinal flora can influence the expression of genes related to cell proliferation and differentiation in the ileum, which is potentially important in promoting the growth and development of pigeons.
Conclusion: Ligilactobacillus salivarius S10 possesses the potential to be used as a probiotic for pigeons, which can influence the expression of gut development-related DEGs by regulating the intestinal flora, and further improve the growth performance of pigeons. This research provides a scientific foundation for developing pigeon-specific probiotics and promotes healthy farming practices for meat pigeons. Furthermore, it opens new avenues for improving the economic efficiency of pigeon farming.
Keywords: Ligilactobacillus salivarius; intestinal flora; intestinal transcriptomics; pigeon; production performance.
Copyright © 2025 Zhao, Li, Yang, Xiao, Zhang, Hong, Ni, Xia, Zhan, Yang and Zhang.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures
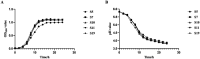

Similar articles
-
Probiotic potential of LAB strains from native Kashmir Flavorella chickens of northwestern Himalayas: A focus on safety and gut health.Res Vet Sci. 2025 Aug;192:105698. doi: 10.1016/j.rvsc.2025.105698. Epub 2025 May 18. Res Vet Sci. 2025. PMID: 40412344
-
Treatment and prevention of pigeon diarrhea through the application of Lactobacillus SNK-6.Poult Sci. 2024 Apr;103(4):103476. doi: 10.1016/j.psj.2024.103476. Epub 2024 Jan 23. Poult Sci. 2024. PMID: 38401224 Free PMC article.
-
Dual bacteriocin and extracellular vesicle-mediated inhibition of Campylobacter jejuni by the potential probiotic candidate Ligilactobacillus salivarius UO.C249.Appl Environ Microbiol. 2024 Aug 21;90(8):e0084524. doi: 10.1128/aem.00845-24. Epub 2024 Jul 30. Appl Environ Microbiol. 2024. PMID: 39078127 Free PMC article.
-
Understanding Ligilactobacillus salivarius from Probiotic Properties to Omics Technology: A Review.Foods. 2024 Mar 15;13(6):895. doi: 10.3390/foods13060895. Foods. 2024. PMID: 38540885 Free PMC article. Review.
-
Ligilactobacillus salivarius functionalities, applications, and manufacturing challenges.Appl Microbiol Biotechnol. 2022 Jan;106(1):57-80. doi: 10.1007/s00253-021-11694-0. Epub 2021 Dec 10. Appl Microbiol Biotechnol. 2022. PMID: 34889985 Review.
References
-
- Agriopoulou S., Stamatelopoulou E., Sachadyn-Król M., Varzakas T. (2020). Lactic acid Bacteria as antibacterial agents to extend the shelf life of fresh and minimally processed fruits and vegetables: quality and safety aspects. Microorganisms 8:952. doi: 10.3390/microorganisms8060952, PMID: - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

