Antifungal activity and mechanism of novel peptide Glycine max antimicrobial peptide (GmAMP) against fluconazole-resistant Candida tropicalis
- PMID: 40416617
- PMCID: PMC12101439
- DOI: 10.7717/peerj.19372
Antifungal activity and mechanism of novel peptide Glycine max antimicrobial peptide (GmAMP) against fluconazole-resistant Candida tropicalis
Abstract
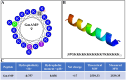
Background: There is a pressing need to create innovative alternative treatment approaches considering the overuse of antifungal drugs causes the number of clinically isolated fluconazole-resistant Candida species to increase. Glycine max antimicrobial peptide (GmAMP) is a novel peptide screened by us using artificial intelligence modeling techniques, and pre-tests showed its strong antimicrobial activity against clinically fluconazole-resistant Candida tropicalis.
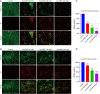
Methods: The study aimed to comprehensively investigate the antimicrobial activity and mechanisms of GmAMP against fluconazole-resistant C. tropicalis. The antifungal activity of GmAMP against fluconazole-resistant C. tropicalis was assessed by using broth microdilution method, growth and fungicidal kinetics, hypha transformation, and antibiofilm assay. To further uncover the potential mechanisms of action of GmAMP, we performed scanning electron microscopy, flow cytometry, cell membrane potential probe 3, 3'-Dipropylthiadicarbocyanine Iodide (DiSC3(5)), and reactive oxygen species (ROS) probe 2', 7'-Dichlorodihydrofluorescein diacetate (DCFH-DA) detection to assess the cellular morphology and structure, membrane permeability, membrane depolarization, and ROS accumulation, respectively. At the same time, we used cytotoxicity and degree of erythrocyte hemolysis assays to assess GmAMP's toxicity in vitro. Cytotoxicity and treatment efficacy were evaluated in vivo by utilizing the Galleria mellonella larvae infection model.
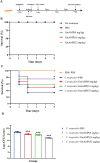
Results: GmAMP exhibited significant antifungal activity against fluconazole-resistant C. tropicalis with a minimum inhibitory concentration (MIC) of 25 µM and demonstrated fungicidal effects at 100 µM within 2 h. GmAMP prevented the transition from yeast to hypha morphology, inhibited the biofilm formation rate of 88.32%, and eradicated the mature biofilm rate of 58.28%. Additionally, GmAMP treatment at 100 µM caused cell structure damage in fluconazole-resistant C. tropicalis, whereas GmAMP treatment at concentrations ranging from 25 to 100 µM caused membrane permeability, depolarization of cell membrane potential, and intracellular ROS accumulation. Moreover, GmAMP enhanced the survival rate of 75% for G. mellonella with fluconazole-resistant C. tropicalis infection as well as reduced fungal burden in vivo by approximately 1.0 × 102 colony forming units per larva (CFU per larva).
Conclusion: GmAMP can disrupt the cell membrane of fluconazole-resistant C. tropicalis and also shows favorable safety and therapeutic efficacy in vivo. Accordingly, GmAMP has the potential to be an agent against drug-resistant fungi.
Keywords: Antifungal activity; Antimicrobial peptide; Candida tropicalis; Drug-resistance; GmAMP.
© 2025 Cai et al.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures



References
-
- Aguiar FLL, Santos NC, de Paula CCS, Andreu D, Baptista GR, Gonçalves S. Antibiofilm activity on Candida albicans and mechanism of action on biomembrane models of the antimicrobial peptide Ctn[15–34] International Journal of Molecular Sciences. 2020;21(21):8339. doi: 10.3390/ijms21218339. - DOI - PMC - PubMed
-
- Alfaro-Vargas P, Bastos-Salas A, Muñoz-Arrieta R, Pereira-Reyes R, Redondo-Solano M, Fernández J, Mora-Villalobos A, López-Gómez JP. Peptaibol production and characterization from Trichoderma asperellum and their action as biofungicide. Journal of Fungi. 2022;8(10):1037. doi: 10.3390/jof8101037. - DOI - PMC - PubMed
-
- Badiee P, Boekhout T, Haddadi P, Mohammadi R, Ghadimi-Moghadam A, Soltani J, Zarei Mahmoudabadi A, Ayatollahi Mousavi SA, Najafzadeh MJ, Diba K, Salimi-Khorashad AR, Amin Shahidi M, Ghasemi F, Jafarian H. Epidemiology and antifungal susceptibility of candida species isolated from 10 tertiary care hospitals in Iran. Microbiology Spectrum. 2022;10(6):e0245322. doi: 10.1128/spectrum.02453-22. - DOI - PMC - PubMed
-
- Bezerra LP, Freitas CDT, Silva AFB, Amaral JL, Neto NAS, Silva RGG, Parra ALC, Goldman GH, Oliveira JTA, Mesquita FP, Souza PFN. Synergistic antifungal activity of synthetic peptides and antifungal drugs against Candida albicans and C. parapsilosis biofilms. Antibiotics. 2022;11(5):553. doi: 10.3390/antibiotics11050553. - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

