Phytochrome E Plays a Role in the Suppression of Germination in Far-Red Light in Tomato
- PMID: 40417128
- PMCID: PMC12100499
- DOI: 10.1002/pld3.70079
Phytochrome E Plays a Role in the Suppression of Germination in Far-Red Light in Tomato
Abstract
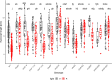
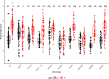
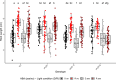
As photoautotrophs, plants use light not only as a source of energy but also as cues for directing growth and development. Phytochromes comprise a small gene family of plant specific light receptors that absorb mostly in the red/far-red portion of the electromagnetic spectrum. These light receptors are well-studied in the model species Arabidopsis thaliana, yet much less is known about their functions in other species. We have generated CRISPR-induced mutations in SlPHYTOCHROME E (SlPHYE) and SlPHYF, produced higher order mutants, and characterized some of their physiological functions in tomato (Solanum lycopersicum). We report that SlphyE plays a major role in detecting far-red light, repressing germination when light conditions are unfavorable for establishing a new seedling. While SlphyE functions on its own, it also synergistically works with another phytochrome, SlphyB1, which by itself only plays a minor role in germination control. Aside from its role in far-red light detection, SlPhyE is also involved in perceiving red light, leading to the repression of hypocotyl elongation and the promotion of light avoidance growth in the roots. SlPhyF acts synergistically with phyB1 during photomorphogenesis but it is not involved in far-red light detection during germination.
Keywords: CRISPR; germination; photomorphogenesis; phytochrome.
© 2025 The Author(s). Plant Direct published by American Society of Plant Biologists and the Society for Experimental Biology and John Wiley & Sons Ltd.
Figures




Similar articles
-
Phytochrome F mediates red light responsiveness additively with phytochromes B1 and B2 in tomato.Plant Physiol. 2023 Apr 3;191(4):2353-2366. doi: 10.1093/plphys/kiad028. Plant Physiol. 2023. PMID: 36670526 Free PMC article.
-
Tomato phyE Is Required for Shade Avoidance in the Absence of phyB1 and phyB2.Front Plant Sci. 2016 Sep 16;7:1275. doi: 10.3389/fpls.2016.01275. eCollection 2016. Front Plant Sci. 2016. PMID: 27695458 Free PMC article.
-
Physiological interactions of phytochromes A, B1 and B2 in the control of development in tomato.Plant J. 2000 Nov;24(3):345-56. doi: 10.1046/j.1365-313x.2000.00879.x. Plant J. 2000. PMID: 11069708
-
Phytochrome-mediated development in land plants: red light sensing evolves to meet the challenges of changing light environments.Mol Ecol. 2006 Oct;15(12):3483-503. doi: 10.1111/j.1365-294X.2006.03051.x. Mol Ecol. 2006. PMID: 17032252 Review.
-
Shedding (far-red) light on phytochrome mechanisms and responses in land plants.Plant Sci. 2014 Mar;217-218:36-46. doi: 10.1016/j.plantsci.2013.11.013. Epub 2013 Nov 28. Plant Sci. 2014. PMID: 24467894 Review.
References
-
- Alba, R. , Kelmenson P. M., Cordonnier‐Pratt M.‐M., and Pratt L. H.. 2000. “The Phytochrome Gene Family in Tomato and the Rapid Differential Evolution of This Family in Angiosperms.” Molecular Biology and Evolution 17: 362–373. - PubMed
-
- Appenroth, K.‐J. , Lenk G., Goldau L., and Sharma R.. 2006. “Tomato Seed Germination: Regulation of Different Response Modes by Phytochrome B2 and Phytochrome A.” Plant, Cell & Environment 29: 701–709. - PubMed
-
- Briggs, W. R. , and Christie J. M.. 2002. “Phototropins 1 and 2: Versatile Plant Blue‐Light Receptors.” Trends in Plant Science 7: 204–210. - PubMed
LinkOut - more resources
Full Text Sources
Research Materials

