Single-cell transcriptomic analysis identifies a stress response Schwann cell subtype
- PMID: 40417312
- PMCID: PMC12103108
- DOI: 10.1515/med-2025-1186
Single-cell transcriptomic analysis identifies a stress response Schwann cell subtype
Abstract
Background: Peripheral nerve injury can lead to sensory, motor, and autonomic nerve dysfunction, significantly impacting patients' quality of life. Schwann cells (SCs), as key components of the peripheral nervous system, play a crucial role in nerve repair. However, many functionally specialized and flexible SC subtypes remain unidentified. Recent advancements in single-cell transcriptomics have enabled a deeper understanding of SC heterogeneity during peripheral nervous system development.
Methods: In this study, we utilized single-cell transcriptomics to investigate SC heterogeneity in the dorsal root ganglia of both normal and spinal nerve injury mouse models.
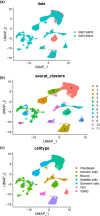
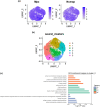
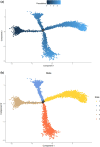
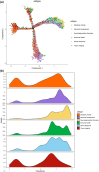
Results: We identified a novel SC subtype associated with pressure sensation, which we termed stress response related SCs (SRSCs). These cells are terminally differentiated and highly express the pressure-sensing gene Npy. Following peripheral nerve injury, SRSCs function as stimulus receptors, receiving external stimuli and transmitting signals to typical repair SCs via the SPP1 signaling network. This interaction promotes dedifferentiation and facilitates injury repair.
Conclusion: Our findings enhance the understanding of SC heterogeneity and reveal SRSCs as key players in nerve repair. These insights provide potential targets for developing novel therapeutic strategies for peripheral nerve diseases.
Keywords: Schwann cell; developmental trajectory; ingle-cell RNA sequencing; peripheral nerve injury; stress response.
© 2025 the author(s), published by De Gruyter.
Conflict of interest statement
Conflict of interest: The authors declare that the study was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures










Similar articles
-
mTORC1 Is Transiently Reactivated in Injured Nerves to Promote c-Jun Elevation and Schwann Cell Dedifferentiation.J Neurosci. 2018 May 16;38(20):4811-4828. doi: 10.1523/JNEUROSCI.3619-17.2018. Epub 2018 Apr 25. J Neurosci. 2018. PMID: 29695414 Free PMC article.
-
Proteomics and transcriptomics of peripheral nerve tissue and cells unravel new aspects of the human Schwann cell repair phenotype.Glia. 2016 Dec;64(12):2133-2153. doi: 10.1002/glia.23045. Epub 2016 Aug 22. Glia. 2016. PMID: 27545331
-
Postinjury Induction of Activated ErbB2 Selectively Hyperactivates Denervated Schwann Cells and Promotes Robust Dorsal Root Axon Regeneration.J Neurosci. 2017 Nov 8;37(45):10955-10970. doi: 10.1523/JNEUROSCI.0903-17.2017. Epub 2017 Oct 5. J Neurosci. 2017. PMID: 28982707 Free PMC article.
-
Schwann cell functions in peripheral nerve development and repair.Neurobiol Dis. 2023 Jan;176:105952. doi: 10.1016/j.nbd.2022.105952. Epub 2022 Dec 7. Neurobiol Dis. 2023. PMID: 36493976 Review.
-
Applications of Proteomics to Nerve Regeneration Research.In: Alzate O, editor. Neuroproteomics. Boca Raton (FL): CRC Press/Taylor & Francis; 2010. Chapter 15. In: Alzate O, editor. Neuroproteomics. Boca Raton (FL): CRC Press/Taylor & Francis; 2010. Chapter 15. PMID: 21882439 Free Books & Documents. Review.
References
-
- Wang ML, Rivlin M, Graham JG, Beredjiklian PK. Peripheral nerve injury, scarring, and recovery. Connect Tissue Res. 2019;60(1):3–9. - PubMed
-
- Li NY, Onor GI, Lemme NJ, Gil JA. Epidemiology of peripheral nerve injuries in sports, exercise, and recreation in the United States, 2009 - 2018. Phys Sportsmed. 2021;49(3):355–62. - PubMed
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous
