Transcriptome and HS-SPME-GC-MS analysis of key genes and flavor components associated with beef marbling
- PMID: 40417364
- PMCID: PMC12098558
- DOI: 10.3389/fvets.2025.1501177
Transcriptome and HS-SPME-GC-MS analysis of key genes and flavor components associated with beef marbling
Abstract
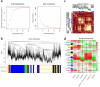
Wagyu cattle are well-known for their rich marbling. Qinchuan cattle have slower-depositing marbling than Wagyu cattle. However, because of an increase in the consumer demand for high-quality beef and the increasingly stringent standards of beef quality, improving the marbling grade of Qinchuan cattle has become particularly crucial. Therefore, we here considered castrated crossbred Wagyu cattle (crossed with Qinchuan cattle) as the research subjects. Flavor substances in the longissimus dorsi muscle (LDM) of A1 and A5 grades were detected through headspace-solid-phase microextraction-gas chromatography-mass spectrometry (HS-SPME-GC-MS) and electronic nose (E-nose) analysis. Fat deposition-regulating functional genes in both groups were identified through RNA sequencing (RNA-seq) and Weighted gene co-expression network analysis (WGCNA). The results showed that the intramuscular fat (IMF) was significantly higher in A5-grade beef (32.96 ± 1.88) than in A1-grade beef (10.91 ± 1.07) (p < 0.01). In total, 41 and 39 flavor compounds were detected in A1 and A5 grade beef, respectively. Seven aroma compounds were identified base on odor activity values (OAVs) ≥ 1, namely decanal, hexanal, nonanal, heptanol, 1-octen-3-ol, pentanol, and hexanoic acid-methyl ester. Additionally, FABP4, PLIN1, LIPE, ACACA, and CIDEA were the key genes primarily involved in cholesterol metabolism, sterol metabolism, and the PPAR signaling pathway in the two grades of beef. This study attempted to offer comprehensive information on marbling formation-associated candidate genes and gene-enriched pathways, which provides data for future research in beef cattle breeding and beef quality improvement.
Keywords: beef grade; crossbred Wagyu cattle; flavor components; marbling; transcriptome.
Copyright © 2025 Ding, Zhang, Zhou, Li, Su, Xu, Qu, Ma, Shi and Kang.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures




References
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

