Association of High Tumor-Stroma Ratio with Prostate Cancer Progression: Insights from Clinical and Genomic Data
- PMID: 40417419
- PMCID: PMC12103176
- DOI: 10.2147/IJGM.S515066
Association of High Tumor-Stroma Ratio with Prostate Cancer Progression: Insights from Clinical and Genomic Data
Abstract
Background: Tumor stroma ratio (TSR) is a prognostic factor in various cancers, but its role in prostate adenocarcinoma (PRAD) remains unclear. This study investigates TSR's prognostic value in PRAD using clinicopathological data, bulk/single-cell RNA sequencing to explore tumor-stroma interactions and identify therapeutic targets.
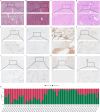
Methods: Two PRAD cohorts (The Cancer Genome Atlas cohort, TCGA; Lanzhou University Second Hospital, LUSH) were analyzed for TSR associations with clinicopathological features and biochemical recurrence (BCR). TSR was assessed via digital image analysis and expert pathologist review. Publicly available bulk/single-cell RNA sequencing data were analyzed to identify TSR-associated genes and predict drug targets, pathways, and immunotherapy responses. Quantitative real-time PCR validated mRNA expression. In vitro assays assessed cell proliferation, growth, and migration, while in vivo xenograft assays validated BGN's role in promoting tumorigenesis.
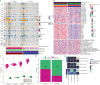
Results: TSR significantly correlated with clinicopathological features (age, Gleason score, stage, seminal vesicle invasion, BCR) in both TCGA (n = 453) and LUSH (n = 320) cohorts. High TSR independently predicted BCR in multivariable Cox regression. High TSR was associated with copy number variations, differentially expressed miRNAs/transcription factors, and metabolic pathways. Predicted anti-cancer drug targets, like Ki8751, showed potential benefit in high-TSR patients. High TSR may correlate with poor immunotherapy response. Notably, downregulation of BGN in cancer-associated fibroblasts (CAFs) significantly suppressed cell proliferation, migration, and invasion in vitro, and in vivo xenograft assays confirmed that BGN downregulation inhibited tumor growth.
Conclusion: This study highlights TSR's prognostic significance in prostate cancer and its association with adverse clinical outcomes and complex tumor-stroma interactions, identifying BGN, a stromal cell-related gene, as a potential therapeutic target for CAFs. However, these findings are limited by the retrospective design, necessitating prospective validation.
Keywords: genomic; prognosis; prostate cancer; single-cell RNA sequencing; tumor-stroma ratio.
© 2025 Xu et al.
Conflict of interest statement
The authors declare that they have no competing interests in this work.
Figures






References
LinkOut - more resources
Full Text Sources
Miscellaneous

