The TP53 Arg72Pro polymorphism predicts visual and neurodegenerative outcomes in retinal detachment
- PMID: 40419469
- PMCID: PMC12106684
- DOI: 10.1038/s41419-025-07739-1
The TP53 Arg72Pro polymorphism predicts visual and neurodegenerative outcomes in retinal detachment
Abstract
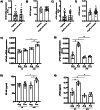
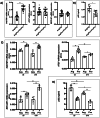
Retinal detachment (RD) separates the retina from the retinal epithelium, causing photoreceptor apoptosis and irreversible vision loss. Even with successful surgical reattachment, complete visual recovery is not guaranteed. The TP53 Arg72Pro polymorphism, implicated in apoptosis, has emerged as a potential predictor of RD outcomes. We investigated the impact of the Arg72Pro polymorphism on retinal neurodegeneration and functional recovery in patients. The underlying mechanisms were analyzed in a humanized TP53 Arg72Pro RD mouse model. In a cohort of 180 patients, carriers of the Pro allele exhibited decreased apoptotic gene expression and improved visual recovery. Complementary findings in mice revealed that the Pro variant preserved photoreceptor integrity and reduced apoptosis rates following RD. Our findings highlight the potential of this TP53 polymorphism as a biomarker for RD outcomes and a tool for tailoring therapies. This study underscores the importance of integrating genetic profiling into personalized medicine approaches to improve recovery of RD patients' visual outcomes.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures





References
-
- Lin JB, Narayanan R, Philippakis E, Yonekawa Y, Apte RS. Retinal detachment. Nat Rev Dis Primers. 2024;10:18. - PubMed
-
- Mitry D, Fleck BW, Wright AF, Campbell H, Charteris DG. Pathogenesis of rhegmatogenous retinal detachment: predisposing anatomy and cell biology. Retina. 2010;30:1561–72. - PubMed
-
- Mitry D, Awan MA, Borooah S, Syrogiannis A, Lim-Fat C, Campbell H, et al. Long-term visual acuity and the duration of macular detachment: findings from a prospective population-based study. Br J Ophthalmol. 2013;97:149–52. - PubMed
-
- Pastor JC, Rojas J, Pastor-Idoate S, Di Lauro S, Gonzalez-Buendia L, Delgado-Tirado S. Proliferative vitreoretinopathy: a new concept of disease pathogenesis and practical consequences. Prog Retin Eye Res. 2016;51:125–55. - PubMed
MeSH terms
Substances
Grants and funding
- RD21/0002/0017, PI21/00727 and RD21/0006/0005/Ministry of Economy and Competitiveness | Instituto de Salud Carlos III (Institute of Health Carlos III)
- RD21/0002/0017 and RD24/0007/0008/Ministry of Economy and Competitiveness | Instituto de Salud Carlos III (Institute of Health Carlos III)
- RD21/0002/0017, PI21/00727 and RD21/0006/0005/Ministry of Economy and Competitiveness | Instituto de Salud Carlos III (Institute of Health Carlos III)
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

