Generating dermatopathology reports from gigapixel whole slide images with HistoGPT
- PMID: 40419470
- PMCID: PMC12106639
- DOI: 10.1038/s41467-025-60014-x
Generating dermatopathology reports from gigapixel whole slide images with HistoGPT
Abstract
Histopathology is the reference standard for diagnosing the presence and nature of many diseases, including cancer. However, analyzing tissue samples under a microscope and summarizing the findings in a comprehensive pathology report is time-consuming, labor-intensive, and non-standardized. To address this problem, we present HistoGPT, a vision language model that generates pathology reports from a patient's multiple full-resolution histology images. It is trained on 15,129 whole slide images from 6705 dermatology patients with corresponding pathology reports. The generated reports match the quality of human-written reports for common and homogeneous malignancies, as confirmed by natural language processing metrics and domain expert analysis. We evaluate HistoGPT in an international, multi-center clinical study and show that it can accurately predict tumor subtypes, tumor thickness, and tumor margins in a zero-shot fashion. Our model demonstrates the potential of artificial intelligence to assist pathologists in evaluating, reporting, and understanding routine dermatopathology cases.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: M.T. is employed by Roche Diagnostics GmbH but conducted his research independently of his work at Roche Diagnostics GmbH as a guest scientist at Helmholtz Munich (Helmholtz Zentrum München—Deutsches Forschungszentrum für Gesundheit und Umwelt GmbH). The remaining authors declare no competing interests. Ethics: An interdisciplinary team of computer scientists, dermatologists, and pathologists from different institutions worked closely together. They shared their expertise and maintained the integrity of the scientific record throughout the study. Local researchers were involved in the research process to ensure that the study was locally relevant. Roles and responsibilities were agreed prior to the study and capacity-building plans were discussed. All research procedures were conducted in accordance with the Declaration of Helsinki. Ethics approval was granted by the Ethics Committee of the Technical University Munich (reference number 2024-98-S-CB) and the Ethics Committee of Westfalen-Lippe (reference number 2024-157-b-S).
Figures

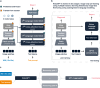



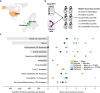
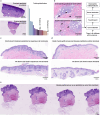
References
-
- Histopathology is ripe for automation. Nat. Biomed. Eng.1, 925 https://www.nature.com/articles/s41551-017-0179-5 (2017). - PubMed
-
- Krug, E. & Varghese, C. Guide for Establishing a Pathology Laboratory in the Context Of Cancer Control (World Health Organization, Geneva, Switzerland, 2020).
-
- Brown, L. Improving histopathology turnaround time: a process management approach. Curr. Diagn. Pathol.10, 444–452 (2004).
-
- van der Laak, J., Litjens, G. & Ciompi, F. Deep learning in histopathology: the path to the clinic. Nat. Med.27, 775–784 (2021). - PubMed
Publication types
MeSH terms
Grants and funding
- 866411/EC | EU Framework Programme for Research and Innovation H2020 | H2020 Priority Excellent Science | H2020 European Research Council (H2020 Excellent Science - European Research Council)
- 101113551/EC | EU Framework Programme for Research and Innovation H2020 | H2020 Priority Excellent Science | H2020 European Research Council (H2020 Excellent Science - European Research Council)
LinkOut - more resources
Full Text Sources
Medical

