Provoking tumor disulfidptosis by single-atom nanozyme via regulating cellular energy supply and reducing power
- PMID: 40419525
- PMCID: PMC12106855
- DOI: 10.1038/s41467-025-60015-w
Provoking tumor disulfidptosis by single-atom nanozyme via regulating cellular energy supply and reducing power
Abstract
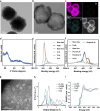
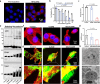
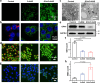
Disulfidptosis, a recently identified form of programmed cell death, is initiated by depletion of endogenous nicotinamide adenine dinucleotide phosphate (NADPH) under glucose starvation. Tumor cells, owing to their heightened requirements of energy and nutrients, are more susceptible to disulfidptosis than normal cells. Here, we introduced an effective strategy to induce tumor disulfidptosis via interrupting cellular energy supply and reducing power by integrating a copper single-atom nanozyme (CuSAE) and glucose oxidase (GOx). GOx induces glucose starvation, impeding generation of NADPH through pentose phosphate pathway (PPP). CuSAE mimics NADPH oxidase, depleting existing NADPH, which intensifies the blockade of disulfide reduction and efficiently triggers disulfidptosis of tumor cells. Furthermore, CuSAE exhibits peroxidase- and glutathione oxidase-mimicking activities, catalyzing generation of •OH radical and depletion cellular GSH, which enhances oxidative stress and exacerbates cell damage. Disulfidptosis is confirmed as the predominant type of cell death induced by GOx/CuSAE. In vivo assays demonstrated the high antitumor potency of GOx/CuSAE in treating with female tumor-bearing mice, with minimal systemic toxicity observed. This work introduces a promising strategy for designing antitumor agents by inducing disulfidptosis. The enzyme hybrids that combine nanozymes and natural enzymes offer a feasible approach to achieve this multifaceted therapeutic goal.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures






References
-
- Carelli, S. et al. Cysteine and glutathione secretion in response to protein disulfide bond formation in the ER. Science277, 1681–1684 (1997). - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

