Characteristic gene expression profile of intestinal mucosa early in life promotes bacterial colonization leading to healthy development of the intestinal environment
- PMID: 40419682
- PMCID: PMC12106659
- DOI: 10.1038/s41598-025-03661-w
Characteristic gene expression profile of intestinal mucosa early in life promotes bacterial colonization leading to healthy development of the intestinal environment
Abstract
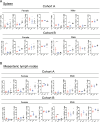
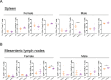
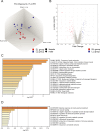
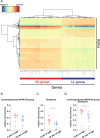
The gut microbiome early in life plays a crucial role in development of the host and affects health throughout life. The definition of a healthy microbiome early in life has not been established, and the underlying mechanism of how a young host selects appropriate microbes for colonization remains unclear. Understanding the mechanism may provide insights into novel preventive and therapeutic strategies by correcting dysbiosis early in life. We employed germ-free mice early in life (4 weeks of age) and later in life (10 weeks of age) for fecal microbiota transfer (FMT) from specific pathogen-free mice. We performed age-unmatched FMT between recipients early in life and donors early or later in life, in addition to common age-matched FMT. Age-matched FMT resulted in significantly different bacterial compositions between recipients early vs. later in life. When the gut microbiome from donors early or later in life was transferred to recipients early in life, bacterial compositions of recipients from donors later in life were similar to those of recipients from donors early in life. This finding suggests that the host early in life has mechanisms to select microbes appropriate for age from the exposed microbiome. We hypothesized that the age-specific intestinal environment promotes age-appropriate intestinal microbiome colonization and examined gene expression in the intestinal mucosa of germ-free mice. We observed that gene expression profiles were different between early vs. later in life. Correlation analysis demonstrated that genera Lachnospiraceae NK4A136 group and Roseburia were positively correlated to genes expressed predominantly early in life, but negatively with genes expressed predominantly later in life. We confirmed that the relative abundance of these genera was significantly higher in specific pathogen-free mice early in life compared with mice later in life. The characteristic gene expression of the intestinal mucosa early in life might play roles in selecting specific bacteria in the intestinal microbiome early in life.
Keywords: Aging; Bacterial colonization; Early life; Gut microbiome; Intestinal gene expression.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Competing interests: The authors declare no competing interests.
Figures


References
-
- Bibbò, S. et al. The role of diet on gut microbiota composition. Eur. Rev. Med. Pharmacol. Sci.20(22), 4742–4749 (2016). - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

