Skeletal muscle endothelial dysfunction through the activin A-PGC1α axis drives progression of cancer cachexia
- PMID: 40419762
- PMCID: PMC12254930
- DOI: 10.1038/s43018-025-00975-6
Skeletal muscle endothelial dysfunction through the activin A-PGC1α axis drives progression of cancer cachexia
Abstract
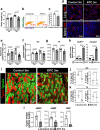
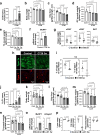
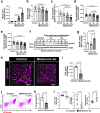
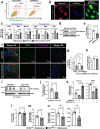
Cachexia is the wasting of skeletal muscle in cancer and is a major complication that impacts a person's quality of life. We hypothesized that cachexia is mediated by dysfunction of the vascular system, which is essential for maintaining perfusion and tempering inappropriate immune responses. Using transparent tissue topography, we discovered that loss of muscle vascular density precedes muscle wasting in multiple complementary tumor models, including pancreatic adenocarcinoma, colon carcinoma, lung adenocarcinoma and melanoma models. We also observed that persons suffering from cancer cachexia exhibit substantial loss of muscle vascular density. As tumors progress, increased circulating activin A remotely suppresses the expression of peroxisome proliferator-activated receptor-γ coactivator 1α (PGC1α) in the muscle endothelium, thus inducing vascular leakage. Restoring endothelial PGC1α activity preserved vascular density and muscle mass in tumor-bearing mice. Our study suggests that restoring muscle endothelial function could be a valuable therapeutic approach for cancer cachexia.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures













References
-
- Biswas, A. K. & Acharyya, S. Cancer-associated cachexia.: a systemic consequence of cancer progression. Annu. Rev. Canc. Biol.4, 391–411 (2020).
-
- Baracos, V. E., Martin, L., Korc, M., Guttridge, D. C. & Fearon, K. C. H. Cancer-associated cachexia. Nat. Rev. Dis. Primers4, 17105 (2018). - PubMed
-
- Argiles, J. M., Stemmler, B., Lopez-Soriano, F. J. & Busquets, S. Inter-tissue communication in cancer cachexia. Nat. Rev. Endocrinol.15, 9–20 (2018). - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials

