Dynamic regulation of integrin β1 phosphorylation supports invasion of breast cancer cells
- PMID: 40419795
- PMCID: PMC12173946
- DOI: 10.1038/s41556-025-01663-4
Dynamic regulation of integrin β1 phosphorylation supports invasion of breast cancer cells
Abstract
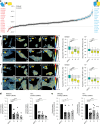
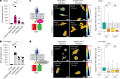
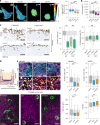
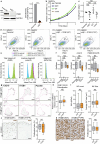
Integrins provide an essential bridge between cancer cells and the extracellular matrix, playing a central role in every stage of disease progression. Despite the recognized importance of integrin phosphorylation in several biological processes, the regulatory mechanisms and their relevance remained elusive. Here we engineer a fluorescence resonance energy transfer biosensor for integrin β1 phosphorylation, screening 96 protein tyrosine phosphatases and identifying Shp2 and PTP-PEST as negative regulators to address this gap. Mutation of the integrin NPxY(783/795) sites revealed the importance of integrin phosphorylation for efficient cancer cell invasion, further supported by inhibition of the identified integrin phosphorylation regulators Shp2 and Src kinase. Using proteomics approaches, we uncovered Cofilin as a component of the phosphorylated integrin-Dok1 complex and linked this axis to effective invadopodia formation, a process supporting breast cancer invasion. These data further implicate dynamic modulation of integrin β1 phosphorylation at NPxY sites at different stages of metastatic dissemination.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures











References
MeSH terms
Substances
Grants and funding
- 346131/Academy of Finland (Suomen Akatemia)
- 841973/EC | EC Seventh Framework Programm | FP7 People: Marie-Curie Actions (FP7-PEOPLE - Specific Programme "People" Implementing the Seventh Framework Programme of the European Community for Research, Technological Development and Demonstration Activities (2007 to 2013))
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

