Structural basis of ubiquitin ligase Nedd4-2 autoinhibition and regulation by calcium and 14-3-3 proteins
- PMID: 40419858
- PMCID: PMC12106849
- DOI: 10.1038/s41467-025-60207-4
Structural basis of ubiquitin ligase Nedd4-2 autoinhibition and regulation by calcium and 14-3-3 proteins
Abstract
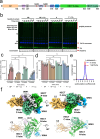
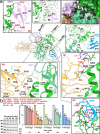
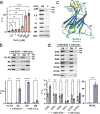
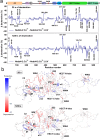
Nedd4-2 E3 ligase regulates Na+ homeostasis by ubiquitinating various channels and membrane transporters, including the epithelial sodium channel ENaC. In turn, Nedd4-2 dysregulation leads to various conditions, including electrolytic imbalance, respiratory distress, hypertension, and kidney diseases. However, Nedd4-2 regulation remains mostly unclear. The present study aims at elucidating Nedd4-2 regulation by structurally characterizing Nedd4-2 and its complexes using several biophysical techniques. Our cryo-EM reconstruction shows that the C2 domain blocks the E2-binding surface of the HECT domain. This blockage, ubiquitin-binding exosite masking by the WW1 domain, catalytic C922 blockage and HECT domain stabilization provide the structural basis for Nedd4-2 autoinhibition. Furthermore, Ca2+-dependent C2 membrane binding disrupts C2/HECT interactions, but not Ca2+ alone, whereas 14-3-3 protein binds to a flexible region of Nedd4-2 containing the WW2 and WW3 domains, thereby inhibiting its catalytic activity and membrane binding. Overall, our data provide key mechanistic insights into Nedd4-2 regulation toward fostering the development of strategies targeting Nedd4-2 function.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures



References
-
- Popovic, D., Vucic, D. & Dikic, I. Ubiquitination in disease pathogenesis and treatment. Nat. Med.20, 1242–1253 (2014). - PubMed
-
- Foot, N., Henshall, T. & Kumar, S. Ubiquitination and the regulation of membrane proteins. Physiol. Rev.97, 253–281 (2017). - PubMed
-
- Rotin, D. & Kumar, S. Physiological functions of the HECT family of ubiquitin ligases. Nat. Rev. Mol. Cell Biol.10, 398–409 (2009). - PubMed
-
- Zheng, N. & Shabek, N. Ubiquitin ligases: structure, function, and regulation. Annu. Rev. Biochem.86, 129–157 (2017). - PubMed
MeSH terms
Substances
Grants and funding
- 23-04686S/Grantová Agentura České Republiky (Grant Agency of the Czech Republic)
- 67985823/Akademie Věd České Republiky (Academy of Sciences of the Czech Republic)
- LM2023042/Ministerstvo Školství, Mládeže a Tělovýchovy (Ministry of Education, Youth and Sports)
- LM2023050/Ministerstvo Školství, Mládeže a Tělovýchovy (Ministry of Education, Youth and Sports)
- 90254/Ministerstvo Školství, Mládeže a Tělovýchovy (Ministry of Education, Youth and Sports)
LinkOut - more resources
Full Text Sources
Miscellaneous

