Identification of single nucleotide polymorphisms (SNPs) associated with heat stress and milk production traits in Chinese holstein cows
- PMID: 40419962
- PMCID: PMC12107732
- DOI: 10.1186/s12864-025-11716-5
Identification of single nucleotide polymorphisms (SNPs) associated with heat stress and milk production traits in Chinese holstein cows
Abstract
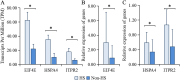
Background: Heat stress (HS) poses a significant challenge to the dairy industry, affecting both the health and productivity of dairy cows. Identifying candidate genes and single nucleotide polymorphisms (SNPs) associated with HS is critical to improving heat tolerance of dairy cows. In our previous work, the eukaryotic translation initiation factor 4E (EIF4E), heat shock protein family A member 4 (HSPA4), and inositol 1,4,5-trisphosphate receptor type 2 (ITPR2) genes were found to play critical roles in the HS response of dairy cows.
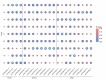
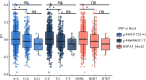
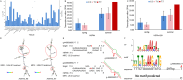
Results: In this study, we further validated the gene expression patterns and genetic effects of the three candidate genes on HS response and milk production in Chinese Holstein cows. A total of 21 SNPs were identified by sequencing the exon and 2000 bp flanking region of the EIF4E (4 SNPs), HSPA4 (8 SNPs), and ITPR2 (9 SNPs) in pooled DNA samples from 70 Holstein bulls. Among these, two SNPs (g.44653172A > G and g.44660065C > T) were located in the coding exon and the 3' untranslated region of the HSPA4 gene, respectively. Association analyses were conducted between identified SNPs and three HS traits and six milk production traits in a population of 1,160 Chinese Holstein cows. The SNP-based association analysis identified significant associations between ten SNPs and HS traits (P < 0.05), as well as eight SNPs and milk production traits (P < 0.05). In the HSPA4 gene, five SNPs (g.44618036G > A, g.44624256 A > C, g.44624428T > C, g.44653172 A > G, and g.44660065 C > T) were significantly associated with rectal temperature (RT; P < 0.05). Notably, no SNPs were associated with milk production traits. Haplotype blocks containing g.44,653,172 A > G and g.44,660,065 C > T also showed significant associations with RT (P < 0.05). Further analysis suggests that g.44,660,065 C > T affects the stability of the mRNA secondary structure, microRNA and transcription factor binding, thereby potentially influencing the gene expression of HSPA4.
Conclusion: In conclusion, we demonstrated that these three genes had significant genetic effects on HS and milk production traits. And g.44,660,065 C > T in the HSPA4 gene could be used as functional molecular markers for the genetic selection of dairy cows for improving heat tolerance without compromising their high milk performance.
Keywords: HSPA4 gene; Association analysis; Dairy cows; Heat stress; Milk production traits.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Ethics approval and consent to participate: This study was conducted in accordance with the guidelines of the Committee on the Ethics of Animal Experimentation at China Agricultural University (Beijing, China), which reviewed and approved all experimental protocols (AW31013202-1-2). No animals were sacrificed or subjected to anesthesia during the course of the study. All animals involved were the property of Beijing Shounong Animal Husbandry Development Co., Ltd., and informed consent was obtained from the farm management prior to the initiation of the study. These procedures also complied with the relevant provisions of the Experimental Animal Management Law of China and conformed to industry standards for animal welfare and management. Consent for publication: Not applicable. Competing interests: The authors declare no competing interests.
Figures


Similar articles
-
Association of Genes TRH, PRL and PRLR with Milk Performance, Reproductive Traits and Heat Stress Response in Dairy Cattle.Int J Mol Sci. 2025 Feb 24;26(5):1963. doi: 10.3390/ijms26051963. Int J Mol Sci. 2025. PMID: 40076589 Free PMC article.
-
Single Nucleotide Polymorphisms of NUCB2 and their Genetic Associations with Milk Production Traits in Dairy Cows.Genes (Basel). 2019 Jun 13;10(6):449. doi: 10.3390/genes10060449. Genes (Basel). 2019. PMID: 31200542 Free PMC article.
-
Single Nucleotide Polymorphisms of ALDH18A1 and MAT2A Genes and Their Genetic Associations with Milk Production Traits of Chinese Holstein Cows.Genes (Basel). 2022 Aug 12;13(8):1437. doi: 10.3390/genes13081437. Genes (Basel). 2022. PMID: 36011348 Free PMC article.
-
Identification of genetic associations and functional SNPs of bovine KLF6 gene on milk production traits in Chinese holstein.BMC Genom Data. 2023 Nov 28;24(1):72. doi: 10.1186/s12863-023-01175-w. BMC Genom Data. 2023. PMID: 38017423 Free PMC article.
-
Unveiling the Genetic Landscape of Feed Efficiency in Holstein Dairy Cows: Insights into Heritability, Genetic Markers, and Pathways via Meta-Analysis.J Anim Sci. 2024 Jan 3;102:skae040. doi: 10.1093/jas/skae040. J Anim Sci. 2024. PMID: 38354297 Free PMC article.
References
-
- FAO, IFAD, UNICEF, WFP, WHO., The State of Food Security and Nutrition in the World 2022. Repurposing food and agricultural policies to make healthy diets more affordable. In. Rome: FAO 2022.
-
- Shephard RW, Maloney SK. A review of thermal stress in cattle. Aust Vet J. 2023;101(11):417–29. - PubMed
-
- Tian H, Wang W, Zheng N, Cheng J, Li S, Zhang Y, Wang J. Identification of diagnostic biomarkers and metabolic pathway shifts of heat-stressed lactating dairy cows. J Proteom. 2015;125:17–28. - PubMed
-
- Kadzere CT, Murphy MR, Silanikove N, Maltz E. Heat stress in lactating dairy cows: a review. Livest Prod Sci. 2002;77(1):59–91.
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

