Comparative genomic analysis of immune-related genes and chemosensory receptors provides insights into the evolution and adaptation of four major domesticated Asian carps
- PMID: 40419972
- PMCID: PMC12105343
- DOI: 10.1186/s12864-025-11719-2
Comparative genomic analysis of immune-related genes and chemosensory receptors provides insights into the evolution and adaptation of four major domesticated Asian carps
Abstract
Background: Ctenopharyngodon idella (grass carp), Mylopharyngodon piceus (black carp), Hypophthalmichthys nobilis (bighead carp), and Hypophthalmichthys molitrix (silver carp), collectively known as the four major domesticated Asian carp, are freshwater fish species from the family Cyprinidae and are widely consumed in China. Current studies on these species primarily focus on immune system regulation and the growth and development of individual species. However, in-depth genomic investigations and comprehensive comparative analysis remained limited.
Methods: The complete genomes of Ctenopharyngodon idella, Mylopharyngodon piceus and Hypophthalmichthys nobilis were assembled using a hybrid approach that integrated both next- and third-generation sequencing reads, followed by annotation using the MAKER2 pipeline. Based on the high-quality genomes of Ctenopharyngodon idella, Mylopharyngodon piceus Hypophthalmichthys nobilis, and Hypophthalmichthys molitrix, a comparative genomic analysis was conducted using bioinformatic tools to investigate gene family evolution in these four domesticated Asian carp species.
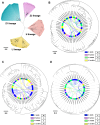
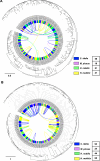
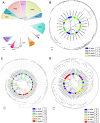
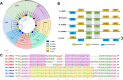
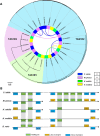
Results: High-quality genomes of Ctenopharyngodon idella, Mylopharyngodon piceus, and Hypophthalmichthys nobilis were assembled, achieving over 90% completeness. Immune-related gene families, including MHC class I and NLRC3-like genes, have undergone rapid evolution, with Ctenopharyngodon idella exhibiting significant expansion of NLRC3-like genes. Massive tandem duplication events were identified in trace amine-associated receptors (TAARs), and rapid expansion was observed in TAAR16 and TAAR29. Additionally, a novel TAAR gene cluster was identified in all four Asian carp species. Comparative genomic analysis revealed the expansion of type 1 taste receptor genes, particularly in Ctenopharyngodon idella and Mylopharyngodon piceus.
Conclusion: This study has successfully constructed the high-quality genomes of Ctenopharyngodon idella, Mylopharyngodon piceus, and Hypophthalmichthys nobilis. The comparative genomic analysis revealed the evolution of immune-related genes and chemosensory receptors in the four major domesticated Asian carp species. These findings suggested the enhanced immunity and sensory perception in these species, providing valuable insights into their adaptation, survival and reproduction.
Keywords: Chemosensory receptors; Comparative genomic analysis; Four major domesticated Asian carp species; Gene family evolution; Immune-related genes.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Ethics approval and consent to participate: Ethical approval does not apply to this study since live animals were not involved. The carp samples were sacrificed at the wet market before purchase. Consent for publication: Not applicable. Competing interests: The authors declare no competing interests.
Figures

References
Publication types
MeSH terms
Substances
Grants and funding
- 14119420/General Research Funds of the Hong Kong Research Grants Council
- 14119420/General Research Funds of the Hong Kong Research Grants Council
- 14119420/General Research Funds of the Hong Kong Research Grants Council
- 14119420/General Research Funds of the Hong Kong Research Grants Council
- 14119420/General Research Funds of the Hong Kong Research Grants Council
- 14119420/General Research Funds of the Hong Kong Research Grants Council
- 14119420/General Research Funds of the Hong Kong Research Grants Council
- 14119420/General Research Funds of the Hong Kong Research Grants Council
- 14119420/General Research Funds of the Hong Kong Research Grants Council
- 14119420/General Research Funds of the Hong Kong Research Grants Council
- T11-709/21-N/Theme-based Research Scheme of the Hong Kong Research Grants Council
- T11-709/21-N/Theme-based Research Scheme of the Hong Kong Research Grants Council
- T11-709/21-N/Theme-based Research Scheme of the Hong Kong Research Grants Council
- T11-709/21-N/Theme-based Research Scheme of the Hong Kong Research Grants Council
- T11-709/21-N/Theme-based Research Scheme of the Hong Kong Research Grants Council
- T11-709/21-N/Theme-based Research Scheme of the Hong Kong Research Grants Council
- T11-709/21-N/Theme-based Research Scheme of the Hong Kong Research Grants Council
- T11-709/21-N/Theme-based Research Scheme of the Hong Kong Research Grants Council
- T11-709/21-N/Theme-based Research Scheme of the Hong Kong Research Grants Council
- T11-709/21-N/Theme-based Research Scheme of the Hong Kong Research Grants Council
LinkOut - more resources
Full Text Sources
Research Materials

