TRIM29 upregulation contributes to chemoresistance in triple negative breast cancer via modulating S100P-β-catenin axis
- PMID: 40420099
- PMCID: PMC12107940
- DOI: 10.1186/s12964-025-02233-9
TRIM29 upregulation contributes to chemoresistance in triple negative breast cancer via modulating S100P-β-catenin axis
Abstract
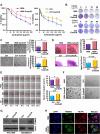
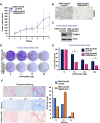
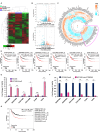
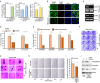
Triple negative breast cancer, an inherently aggressive disease, is further impaired by the limited therapeutic options and chemotherapy-resistance; hence, elucidating the signaling nodes underlying chemotherapy resistance is of major interest. Focusing on the differentially expressed genes in recurrent TNBC, we identified TRIM29, a ubiquitin ligase belonging to TRIM family, as a uniquely enriched protein in chemoresistant TNBC. Here, we demonstrate that chemoresistant TNBC cells are inherently aggressive, exhibiting elevated growth and migration potential compared to chemosensitive cells, and in particular, they possess higher TRIM29 expression whose expression level modulation results in altered chemosensitivity. TRIM29 overexpression reduces chemotherapy response whereas TRIM29 knockout not only increases chemosensitivity but also reduces TNBC tumor growth. Tumor-dissociated cells maintain TRIM29 knockout status as well as exhibit similar functional alterations as chemoresistant TNBC cells. Mechanistically, RNA-sequencing of parental-chemosensitive, chemoresistant-inherently overexpressing TRIM29 and chemoresistant-TRIM29 knockout TNBC cells reveals a unique set of genes (S100P, SERPINB3, SERPINB4, CEACAM5, CEACAM6 and CDH6) showing significant upregulation with the acquisition of chemoresistance and downregulation with the TRIM29 knockout. Furthermore, an enrichment of β-catenin pathway in chemoresistant TNBC cells is observed. We uncovered a functional network where S100P, a metastasis inducing secretory factor, bidirectionally interacts with TRIM29, and modulates the expression of SERPINB3, SERPINB4, CEACAM5, CEACAM6 as well as β-catenin pathway genes. Showing the functional importance, S100P inhibitor reduces the growth and mammosphere formation in chemoresistant TNBC. Moreover, combining β-catenin inhibitor with chemotherapy shows synergistic inhibition of chemoresistant TNBC cells. Indeed, higher expression of TRIM29, S100P and β-catenin associates with reduced recurrence free survival. This work proposes TRIM29 as an important node that modulates a unique gene network in chemoresistant TNBC and whose biological impact is mediated by modulation of S100P and β-catenin.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Ethics approval and consent to participate: No clinical data. All animal studies were approved by JHU ACUC. Consent for publication: All authors consent for publication. Competing interests: The authors declare no competing interests.
Figures



References
-
- Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, Bray F, Global Cancer Statistics 2020.: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries, CA: a Cancer Journal for Clinicians. 2021;71:209–49. - PubMed
-
- Elias AD. Triple-negative breast cancer: a short review. Am J Clin Oncol. 2010;33:637–45. - PubMed
-
- Bai X, Ni J, Beretov J, Graham P, Li Y. Triple-negative breast cancer therapeutic resistance: where is the Achilles’ heel? Cancer Lett. 2021;497:100–11. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

