Prediction of KRAS gene mutations in colorectal cancer using a CT-based radiomic model
- PMID: 40421293
- PMCID: PMC12104245
- DOI: 10.3389/fmed.2025.1592497
Prediction of KRAS gene mutations in colorectal cancer using a CT-based radiomic model
Erratum in
-
Corrigendum: Prediction of KRAS gene mutations in colorectal cancer using a CT-based radiomic model.Front Med (Lausanne). 2025 Jun 24;12:1630007. doi: 10.3389/fmed.2025.1630007. eCollection 2025. Front Med (Lausanne). 2025. PMID: 40630476 Free PMC article.
Abstract
Background: Determining the KRAS gene mutation status in colorectal cancer (CRC) before surgery is highly important for an individualized clinical treatment. This study aimed to explore the clinical value of radiomics models based on CT images in predicting the KRAS mutation status in patients with CRC.
Methods: A total of 201 CRC patients who underwent surgery and pathology examinations from March 2022 to January 2025 were included. They were randomly allocated to a training group (160 patients) or a testing group (41 patients) at a ratio of 8:2. All patients underwent plain CT and contrast-enhanced examinations before surgery. The 3D segmentation of the tumour was manually delineated by two radiologists who were unaware of the pathological results and KRAS gene detection outcomes. The PyRadiomics package in Python was used to extract 2,264 radiomic features from each ROI. After dimensionality reduction, machine learning methods such as extremely randomized trees (ERT), random forest (RF), XGBoost, Bagging, and CatBoost were used for model construction. The performance of the models was compared using the area under the receiver operating characteristic curve (AUC), accuracy, sensitivity, and specificity. The Delong test was employed to assess the differences between the various models.
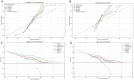
Results: After feature selection, the top 8 features with the highest mutual information scores were extracted to construct a prediction model. The Delong test revealed that the XGBoost model, which is based on CT images from the vein phase, performed the best, with AUC values of 0.90 and 0.81 in the training and test sets, respectively. The calibration curve indicated a high consistency between the actual and predicted probabilities of the samples. The decision curve analysis results revealed that the XGBoost model exhibited the highest net clinical benefit among all the models.
Conclusion: In this study, a highly accurate radiomics model was developed for KRAS gene mutation status prediction in patients with CRC before surgery. This technique avoids the potential risks of tumour rupture and dissemination during biopsy and can serve as a powerful tool to assist doctors in developing personalized and precise targeted treatments for colorectal cancer, which highly important in clinical work.
Keywords: KRAS gene mutation; colorectal cancer; computed tomography; machine learning; radiomics.
Copyright © 2025 Wang, Zhang, Fan, Wang, Le, Ai, Du, Feng and Li.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


References
LinkOut - more resources
Full Text Sources
Miscellaneous

