MDTAP: a tool to analyze permeation events across membrane proteins
- PMID: 40421423
- PMCID: PMC12104522
- DOI: 10.1093/bioadv/vbaf102
MDTAP: a tool to analyze permeation events across membrane proteins
Abstract
Motivation: Molecular dynamics (MD) simulations provide critical insights into the transport of solutes, solvents, and drug molecules across protein channels embedded in a membrane bilayer. However, identifying and analyzing the permeation events from complex simulation data remains as a challenging and laborious task. Thus, an automated tool that facilitates the capture of permeation events of any molecular type across any channel is essential to streamline MD trajectory analysis and enhance the understanding of biological processes in a timely manner.
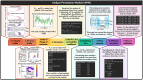
Results: Molecular Dynamics Trajectory Analysis of Permeation (MDTAP) is a Linux/Mac-based software that automatically detects permeation events across membrane-embedded protein and nucleic acid channels. The tool accepts trajectories in DCD (CHARMM/NAMD) and PDB format (obtained from any MD simulation package) and employs bash scripts to analyze the input trajectories to characterize the molecular permeation. The efficiency of MDTAP is demonstrated using MD trajectories of Escherichia coli outer membrane protein Wzi and E. coli Aquaporin Z. MDTAP can also analyze permeation across heterogeneous lipid membranes and artificial nucleic acid channels, addressing their growing importance. Thus, MDTAP simplifies trajectory analysis and also reduces the need for manual inspection.
Availability and implementation: MDTAP is open-source and is freely available on GitHub (https://github.com/MBL-lab/MDTAP), including source code, installation instructions, and usage documentation.
© The Author(s) 2025. Published by Oxford University Press.
Conflict of interest statement
The authors declare that they have no known competing interests.
Figures


References
-
- Niitsu A, Sugita Y. Towards de novo design of transmembrane α-helical assemblies using structural modelling and molecular dynamics simulation. Phys Chem Chem Phys 2023;25:3595–606. - PubMed
-
- Beckstein O, Biggin PC, Bond P et al. Ion channel gating: insights via molecular simulations. FEBS Lett 2003;555:85–90. - PubMed
-
- Carnevale V, Delemotte L, Howard RJ. Molecular dynamics simulations of ion channels. Trends Biochem Sci 2021;46:621–2. - PubMed
LinkOut - more resources
Full Text Sources
Miscellaneous

