SARS-CoV-2 point mutations are over-represented in terminal loops of RNA stem-loop structures that can be resolved by Nsp13 helicase in a unique manner with respect to nucleotide dependence
- PMID: 40421800
- PMCID: PMC12107433
- DOI: 10.1093/nar/gkaf447
SARS-CoV-2 point mutations are over-represented in terminal loops of RNA stem-loop structures that can be resolved by Nsp13 helicase in a unique manner with respect to nucleotide dependence
Abstract
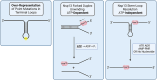
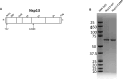
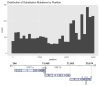
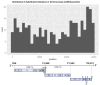
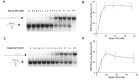
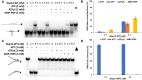
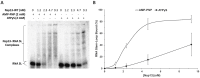
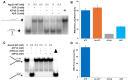
To improve health outcomes for COVID-19 (coronavirus disease 2019) patients, the factors that influence coronavirus genome variation need to be ascertained. The SARS-CoV-2 (severe acute respiratory syndrome coronavirus 2) genome is rich in predicted RNA secondary structures, particularly stem-loops (SLs) formed by intramolecular base pairing within palindromic sequences. We analyzed the NCBI Virus collection of SARS-CoV-2 genome sequences from COVID-19 individuals to map variants relative to SL structural elements. Point mutations in the SARS-CoV-2 genome, with a C-to-U transition bias, were over-represented in unpaired nucleotides and, more specifically, within the terminal loops of RNA SL structures. As the sole helicase encoded by SARS-CoV-2, Nsp13 may operate in the timely resolution of secondary RNA structures to facilitate SARS-CoV-2 RNA copying or processing. We characterized Nsp13 to resolve SARS-CoV-2 sequence-derived unimolecular RNA SL substrates and determined that it does so in a functionally cooperative manner. In addition to ATP, Nsp13 resolves the unimolecular RNA SL structure in the absence of nucleotide, in contrast to the strict ATP requirement for a bimolecular RNA forked duplex. We suggest a model in which a series of binary and ternary complex interactions of Nsp13 with nucleotide and/or RNA SL pose mechanistic implications for RNA SL resolution.
Published by Oxford University Press on behalf of Nucleic Acids Research 2025.
Conflict of interest statement
None declared.
Figures

Similar articles
-
ATPase-dependent duplex nucleic acid unwinding by SARS-CoV-2 nsP13 relies on facile binding and translocation along single-stranded nucleic acid.J Biol Chem. 2025 Jul;301(7):110373. doi: 10.1016/j.jbc.2025.110373. Epub 2025 Jun 12. J Biol Chem. 2025. PMID: 40516869 Free PMC article.
-
SARS-Coronavirus-2 Nsp13 Possesses NTPase and RNA Helicase Activities That Can Be Inhibited by Bismuth Salts.Virol Sin. 2020 Jun;35(3):321-329. doi: 10.1007/s12250-020-00242-1. Epub 2020 Jun 4. Virol Sin. 2020. PMID: 32500504 Free PMC article.
-
The stalk domain of SARS-CoV-2 NSP13 is essential for its helicase activity.Biochem Biophys Res Commun. 2022 Apr 23;601:129-136. doi: 10.1016/j.bbrc.2022.02.068. Epub 2022 Feb 23. Biochem Biophys Res Commun. 2022. PMID: 35245742 Free PMC article.
-
The Coronavirus helicase in replication.Virus Res. 2024 Aug;346:199401. doi: 10.1016/j.virusres.2024.199401. Epub 2024 May 31. Virus Res. 2024. PMID: 38796132 Free PMC article. Review.
-
SARS-CoV ORF1b-encoded nonstructural proteins 12-16: replicative enzymes as antiviral targets.Antiviral Res. 2014 Jan;101:122-30. doi: 10.1016/j.antiviral.2013.11.006. Epub 2013 Nov 20. Antiviral Res. 2014. PMID: 24269475 Free PMC article. Review.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

