Refining Drug-Induced Cholestasis Prediction: An Explainable Consensus Model Integrating Chemical and Biological Fingerprints
- PMID: 40421892
- PMCID: PMC12152943
- DOI: 10.1021/acs.jcim.4c02363
Refining Drug-Induced Cholestasis Prediction: An Explainable Consensus Model Integrating Chemical and Biological Fingerprints
Abstract
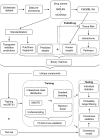
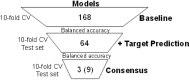
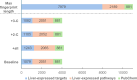
Effective drug safety assessment, guided by the 3R principle (Replacement, Reduction, Refinement) to minimize animal testing, is critical in early drug development. Drug-induced liver injury (DILI), particularly drug-induced cholestasis (DIC), remains a major challenge. This study introduces a computational method for predicting DIC by integrating PubChem substructure fingerprints with biological data from liver-expressed targets and pathways, alongside nine hepatic transporter inhibition models. To address class imbalance in the public cholestasis data set, we employed undersampling, a technique that constructs a small and robust consensus model by evaluating distinct subsets. The most effective baseline model, which combined PubChem substructure fingerprints, pathway data and hepatic transporter inhibition predictions, achieved a Matthews correlation coefficient (MCC) of 0.29 and a sensitivity of 0.79, as validated through 10-fold cross-validation. Subsequently, target prediction using four publicly available tools was employed to enrich the sparse compound-target interaction matrix. Although this approach showed lower sensitivity compared to experimentally derived targets and pathways, it highlighted the value of incorporating specific systems biology related information. Feature importance analysis identified albumin as a potential target linked to cholestasis within our predictive model, suggesting a connection worth further investigation. By employing an expanded consensus model and applying probability range filtering, the refined method achieved an MCC of 0.38 and a sensitivity of 0.80, thereby enhancing decision-making confidence. This approach advances DIC prediction by integrating biological and chemical descriptors, offering a reliable and explainable model.
Figures


Similar articles
-
A Change in Bile Flow: Looking Beyond Transporter Inhibition in the Development of Drug-induced Cholestasis.Curr Drug Metab. 2019;20(8):621-632. doi: 10.2174/1389200220666190709170256. Curr Drug Metab. 2019. PMID: 31288715 Free PMC article. Review.
-
Identification of reversible OATP1B1 and time-dependent CYP3A4 inhibition as the major risk factors for drug-induced cholestasis (DIC).Arch Toxicol. 2024 Oct;98(10):3409-3424. doi: 10.1007/s00204-024-03794-3. Epub 2024 Jul 18. Arch Toxicol. 2024. PMID: 39023798
-
Predicting Drug-Induced Cholestasis with the Help of Hepatic Transporters-An in Silico Modeling Approach.J Chem Inf Model. 2017 Mar 27;57(3):608-615. doi: 10.1021/acs.jcim.6b00518. Epub 2017 Mar 8. J Chem Inf Model. 2017. PMID: 28166633 Free PMC article.
-
Unraveling the mechanisms underlying drug-induced cholestatic liver injury: identifying key genes using machine learning techniques on human in vitro data sets.Arch Toxicol. 2023 Nov;97(11):2969-2981. doi: 10.1007/s00204-023-03583-4. Epub 2023 Aug 21. Arch Toxicol. 2023. PMID: 37603094 Free PMC article.
-
Advances in drug-induced cholestasis: Clinical perspectives, potential mechanisms and in vitro systems.Food Chem Toxicol. 2018 Oct;120:196-212. doi: 10.1016/j.fct.2018.07.017. Epub 2018 Jul 7. Food Chem Toxicol. 2018. PMID: 29990576 Review.
References
-
- Singh N., Vayer P., Tanwar S., Poyet J., Tsaioun K., Villoutreix B. O.. Drug Discovery and Development: Introduction to the General Public and Patient Groups. Front. Drug Discovery. 2023;3:1–11. doi: 10.3389/fddsv.2023.1201419. - DOI
-
- Russell, W. M. S. ; Burch, R. L. . The Principles of Humane Experimental Technique; Methuen & Co. Ltd: London, 1959.
-
- S.5002 - 117th Congress (2021–2022): FDA Modernization Act 2.0; Library of Congress, 2022. https://www.congress.gov/bill/117th-congress/senate-bill/5002. (accessed 23 August 2024).
MeSH terms
LinkOut - more resources
Full Text Sources

