One-Pot Detection of miRNA by Dual Rolling Circle Amplification at Ambient Temperature with High Specificity and Sensitivity
- PMID: 40422056
- PMCID: PMC12109915
- DOI: 10.3390/bios15050317
One-Pot Detection of miRNA by Dual Rolling Circle Amplification at Ambient Temperature with High Specificity and Sensitivity
Abstract
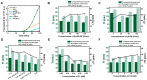
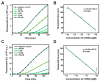
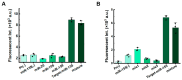
Rolling circle amplification (RCA) at ambient temperature is prone to false positive signals during nucleic acid detection, which makes it challenging to establish an efficient RCA detection method. The false positive signals are primarily caused by binding of non-target nucleic acids to the circular single-stranded template, leading to non-specific amplification. Here, we present an RCA method for miRNA detection at 37 °C using two circular ssDNAs, each of which is formed by ligating the intramolecularly formed nick (without any splint) in a secondary structure. The specific target recognition is realized by utilizing low concentrations (0.1 nM) of circular ssDNA1 (C1). A phosphorothioate modification is present at G*AATTC on C1 to generate a nick for primer extension during the primer self-generated rolling circle amplification (PG-RCA). The fragmented amplification products are used as primers for the following RCA that serves as signal amplification using circular ssDNA2 (C2). Notably, the absence of splints and the low concentration of C1 significantly inhibits non-target binding, thus minimizing false positive signals. A high concentration (10 nM) of C2 is used to carry out linear rolling circle amplification (LRCA), which is highly specific. This strategy demonstrates a good linear response to 0.01-100 pM of miRNA with a detection limit of 7.76 fM (miR-155). Moreover, it can distinguish single-nucleotide mismatch in the target miRNA, enabling the rapid one-pot detection of miRNA at 37 °C. Accordingly, this method performs with high specificity and sensitivity. This approach is suitable for clinical serum sample analysis and offers a strategy for developing specific biosensors and diagnostic tools.
Keywords: circular ssDNA; dual RCA; microRNA detection; splint; ssDNA circularization.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures


Similar articles
-
A dual-signal amplification strategy based on rolling circle amplification and APE1-assisted amplification for highly sensitive and specific miRNA analysis for early diagnosis of alzheimer's disease.Talanta. 2024 May 15;272:125747. doi: 10.1016/j.talanta.2024.125747. Epub 2024 Feb 10. Talanta. 2024. PMID: 38364557
-
Rapid and ultrasensitive miRNA detection by combining endonuclease reactions in a rolling circle amplification (RCA)-based hairpin DNA fluorescent assay.Anal Bioanal Chem. 2023 Apr;415(10):1991-1999. doi: 10.1007/s00216-023-04618-6. Epub 2023 Feb 28. Anal Bioanal Chem. 2023. PMID: 36853410
-
Nicking-enhanced rolling circle amplification for sensitive fluorescent detection of cancer-related microRNAs.Anal Bioanal Chem. 2018 Oct;410(26):6819-6826. doi: 10.1007/s00216-018-1277-2. Epub 2018 Aug 1. Anal Bioanal Chem. 2018. PMID: 30066196
-
Recent advances in rolling circle amplification-based biosensing strategies-A review.Anal Chim Acta. 2021 Mar 1;1148:238187. doi: 10.1016/j.aca.2020.12.062. Epub 2020 Dec 31. Anal Chim Acta. 2021. PMID: 33516384 Review.
-
Recent advances in biological detection with rolling circle amplification: design strategy, biosensing mechanism, and practical applications.Analyst. 2022 Jul 22;147(15):3396-3414. doi: 10.1039/d2an00556e. Analyst. 2022. PMID: 35748818 Review.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

