The sulfur assimilation pathway mitigates redox stress from acidic pH in Salmonella Typhi H58
- PMID: 40422406
- PMCID: PMC12239559
- DOI: 10.1128/mbio.00467-25
The sulfur assimilation pathway mitigates redox stress from acidic pH in Salmonella Typhi H58
Abstract
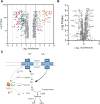
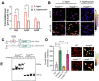
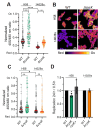
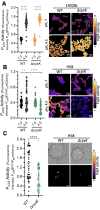
Salmonella enterica serovar Typhi is the causative agent of typhoid fever, a human-restricted systemic infection. The rapidly disseminating multi-drug resistant H58 subclade is endemic in Africa, Asia, and Oceania, yet little is known regarding its intracellular behavior and virulence properties. It was of interest to understand the H58 response to host cell infection in terms of its response to acid stress and subsequent changes in gene regulation. We compared the H58 response in vitro and during infection of THP-1 human macrophages with the well-characterized response of Salmonella Typhimurium, which causes gastroenteritis. In S. Typhimurium infections, bacteria reside in an acidic intracellular vacuole and become acidified, driving the expression of pathogenicity island 2 genes (SPI-2). SPI-2 virulence factors modify the vacuole and enable bacterial replication. In response to acid stress, the sulfur assimilation pathway is highly upregulated and unique to H58. Replacing the Typhi cysK promoter with the Typhimurium promoter resulted in a cysK gene that was upregulated by acid stress in the H58 background, highlighting the differential regulation between the two serovars. In acidic conditions, H58 experienced much greater redox stress compared with S. Typhimurium, and the sulfur assimilation pathway was required to mitigate the redox stress. Higher redox stress modified the transcriptional regulator SsrB, resulting in diminished secretion of the SPI-2 virulence factor SifA. Our results highlight significant differences between S. Typhi H58 and S. Typhimurium and emphasize the importance of studying S. Typhi strains directly to understand their unique behavior during pathogenesis.
Importance: In this study, we examined the clinically relevant, multi-drug resistant Salmonella Typhi strain H58, which is rapidly disseminating across Southeast Asia, Africa, and Oceania. It has heretofore been uncharacterized in terms of its gene regulation. Using human THP-1 macrophages, we discovered that S. Typhi strongly activates the sulfur utilization pathway in response to acid stress encountered in the vacuole once Typhi is inside host cells. Our novel findings were that S. Typhi experiences substantially higher redox stress compared with Typhimurium, and it requires the sulfur utilization pathway to mitigate this stress. This pathway is not upregulated in Typhimurium and represents a divergence in the response of these two serovars. We emphasize that S. Typhimurium is not a reasonable model for understanding H58, a serovar that is seriously impacting human health.
Keywords: H58; Salmonella Typhi; Salmonella pathogenicity island 2; SifA; SsrB; acid stress response; redox; sulfur assimilation.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Similar articles
-
Propionylation of Fis K32 in Salmonella enterica serovar Typhi: a key modification affecting pathogenicity.Future Microbiol. 2025 Mar;20(4):295-303. doi: 10.1080/17460913.2025.2460338. Epub 2025 Jan 30. Future Microbiol. 2025. PMID: 39885648
-
Salmonella exploits host- and bacterial-derived β-alanine for replication inside host macrophages.Elife. 2025 Jun 19;13:RP103714. doi: 10.7554/eLife.103714. Elife. 2025. PMID: 40536105 Free PMC article.
-
Distinct adaptation and epidemiological success of different genotypes within Salmonella enterica serovar Dublin.Elife. 2025 Jun 25;13:RP102253. doi: 10.7554/eLife.102253. Elife. 2025. PMID: 40560760 Free PMC article.
-
Rapid diagnostic tests for typhoid and paratyphoid (enteric) fever.Cochrane Database Syst Rev. 2017 May 26;5(5):CD008892. doi: 10.1002/14651858.CD008892.pub2. Cochrane Database Syst Rev. 2017. PMID: 28545155 Free PMC article.
-
Vaccines for preventing typhoid fever.Cochrane Database Syst Rev. 2018 May 31;5(5):CD001261. doi: 10.1002/14651858.CD001261.pub4. Cochrane Database Syst Rev. 2018. PMID: 29851031 Free PMC article.
References
-
- Stanaway JD, Reiner RC, Blacker BF, Goldberg EM, Khalil IA, Troeger CE, Andrews JR, Bhutta ZA, Crump JA, Im J, et al. 2019. The global burden of typhoid and paratyphoid fevers: a systematic analysis for the global burden of disease study 2017. Lancet Infect Dis 19:369–381. doi: 10.1016/S1473-3099(18)30685-6 - DOI - PMC - PubMed
-
- Wong VK, Baker S, Connor TR, Pickard D, Page AJ, Dave J, Murphy N, Holliman R, Sefton A, Millar M, Dyson ZA, Dougan G, Holt KE, International Typhoid C. 2016. An extended genotyping framework for Salmonella enterica serovar Typhi, the cause of human typhoid. Nat Commun 7:12827. doi: 10.1038/ncomms12827 - DOI - PMC - PubMed
-
- Desai SK, Zhou Y, Dilawari R, Routh AL, Popov V, Kenney LJ. 2023. RpoS activates Salmonella Typhi biofilms and drives persistence in a Zebrafish model. bioRxiv. doi: 10.1101/2023.10.26.564249 - DOI
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

