Integrating Dynamical Systems Modeling with Spatiotemporal scRNA-Seq Data Analysis
- PMID: 40422408
- PMCID: PMC12109813
- DOI: 10.3390/e27050453
Integrating Dynamical Systems Modeling with Spatiotemporal scRNA-Seq Data Analysis
Abstract
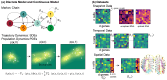
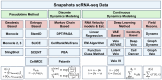
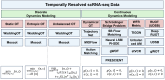
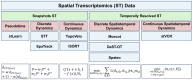
Understanding the dynamic nature of biological systems is fundamental to deciphering cellular behavior, developmental processes, and disease progression. Single-cell RNA sequencing (scRNA-seq) has provided static snapshots of gene expression, offering valuable insights into cellular states at a single time point. Recent advancements in temporally resolved scRNA-seq, spatial transcriptomics (ST), and time-series spatial transcriptomics (temporal-ST) have further revolutionized our ability to study the spatiotemporal dynamics of individual cells. These technologies, when combined with computational frameworks such as Markov chains, stochastic differential equations (SDEs), and generative models like optimal transport and Schrödinger bridges, enable the reconstruction of dynamic cellular trajectories and cell fate decisions. This review discusses how these dynamical system approaches offer new opportunities to model and infer cellular dynamics from a systematic perspective.
Keywords: cellular trajectories; computational modeling; single-cell RNA sequencing; spatiotemporal dynamics.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures
Similar articles
-
Computational solutions for spatial transcriptomics.Comput Struct Biotechnol J. 2022 Sep 1;20:4870-4884. doi: 10.1016/j.csbj.2022.08.043. eCollection 2022. Comput Struct Biotechnol J. 2022. PMID: 36147664 Free PMC article. Review.
-
Modeling single cell trajectory using forward-backward stochastic differential equations.PLoS Comput Biol. 2024 Apr 15;20(4):e1012015. doi: 10.1371/journal.pcbi.1012015. eCollection 2024 Apr. PLoS Comput Biol. 2024. PMID: 38620017 Free PMC article.
-
Multivariate stochastic modeling for transcriptional dynamics with cell-specific latent time using SDEvelo.Nat Commun. 2024 Dec 30;15(1):10849. doi: 10.1038/s41467-024-55146-5. Nat Commun. 2024. PMID: 39738101 Free PMC article.
-
SpaIM: Single-cell Spatial Transcriptomics Imputation via Style Transfer.bioRxiv [Preprint]. 2025 Jan 27:2025.01.24.634756. doi: 10.1101/2025.01.24.634756. bioRxiv. 2025. PMID: 39975319 Free PMC article. Preprint.
-
Single-cell RNA sequencing and spatial transcriptome analysis in bladder cancer: Current status and future perspectives.Bladder Cancer. 2025 Feb 21;11(1):23523735251322017. doi: 10.1177/23523735251322017. eCollection 2025 Jan-Mar. Bladder Cancer. 2025. PMID: 40034247 Free PMC article. Review.
References
-
- Lei J. Mathematical modeling of heterogeneous stem cell regeneration: From cell division to Waddington’s epigenetic landscape. arXiv. 20232309.08064
-
- Schiebinger G. Reconstructing developmental landscapes and trajectories from single-cell data. Curr. Opin. Syst. Biol. 2021;27:100351. doi: 10.1016/j.coisb.2021.06.002. - DOI
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources

