Exploring the Mechanism of Luteolin in Protecting Chickens from Ammonia Poisoning Based on Proteomic Technology
- PMID: 40422902
- PMCID: PMC12113253
- DOI: 10.3390/metabo15050326
Exploring the Mechanism of Luteolin in Protecting Chickens from Ammonia Poisoning Based on Proteomic Technology
Abstract
Background: Ammonia (NH3), a harmful gas, reduces livestock productivity, threatens their health, and causes economic losses. Luteolin (Lut), an anti-inflammatory flavonoid, may counteract these effects.
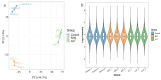
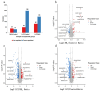
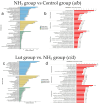
Methods: Our study explored luteolin's protective mechanisms on chicken splenic lymphocytes under ammonia stress using a simulation model and four-dimensional fast data-independent acquisition (4D-FastDIA) proteomics. We identified 316 proteins, with 69 related to ammonia's negative effects and 247 to Lut's protection. Thirty differentially expressed proteins (DEPs) were common to both groups, with 27 showing counter-regulation with Lut.
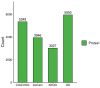
Results: Gene Ontology (GO) analysis showed DEPs enriched in molecular responses to interferons and the negative regulation of immune responses, mainly located extracellularly. Molecular function analysis revealed DEPs in antigen binding and synthase activity. Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis linked DEPs to pathways like estrogen signaling, NOD-like receptor signaling, cytokine-cytokine receptor interaction, and JAK-STAT signaling. The quantitative real-time polymerase chain reaction (qRT-PCR) results indicated that the mRNA levels of Interferon Alpha and Beta Receptor subunit 2 (IFNAR2) and Signal Transducer and Activator of Transcription 1 (STAT1) were trending downward. This observation was in strong agreement with the downregulation noted in the proteomics analysis.
Conclusions: Lut's protective role against ammonia's adverse effects on chicken splenic lymphocytes is linked to the modulation of key signaling pathways, offering insights for further research on treating ammonia exposure with Lut.
Keywords: ammonia; anti-inflammatory; apoptosis; chicken; luteolin; proteomics.
Conflict of interest statement
The authors certify that there are no competing interests that could potentially bias the impartiality of this report.
Figures






References
-
- Shah S.W.A., Chen J., Han Q., Xu Y., Ishfaq M., Teng X. Ammonia inhalation impaired immune function and mitochondrial integrity in the broilers bursa of fabricius: Implication of oxidative stress and apoptosis. Ecotoxicol. Environ. Saf. 2020;190:110078. doi: 10.1016/j.ecoenv.2019.110078. - DOI - PubMed
-
- Chen D., Ning F., Zhang J., Tang Y., Teng X. NF-κB pathway took part in the development of apoptosis mediated by miR-15a and oxidative stress via mitochondrial pathway in ammonia-treated chicken splenic lymphocytes. Sci. Total Environ. 2020;729:139017. doi: 10.1016/j.scitotenv.2020.139017. - DOI - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

