Gated Nanosensor for Sulphate-Reducing Bacteria Detection
- PMID: 40423163
- PMCID: PMC12114299
- DOI: 10.3390/nano15100774
Gated Nanosensor for Sulphate-Reducing Bacteria Detection
Abstract
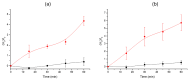
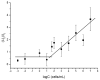
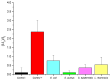
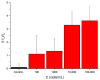
Desulfovibrio vulgaris is an anaerobic microorganism belonging to the group of sulphate-reducing bacteria (SRB). SRB form biofilms on metal surfaces in water supply networks, producing a microbiologically influenced corrosion (MIC). This process produces the deterioration of metal surfaces, leading to high economic costs and different environmental safety and health problems related to its chemical treatment. For that reason, rapid and accurate detection methods of SRB are needed. In this work, a new detection system for Desulfovibrio has been developed using gated nanoporous materials. The probe is based on hybrid nanoporous alumina films encapsulating a fluorescent molecule (rhodamine B), whose release is controlled by an oligonucleotide gate. Upon exposure to Desulfovibrio's genomic material, a movement of the oligonucleotide gatekeeper happens, resulting in the selective delivery of the entrapped rhodamine B. The developed material shows high selectivity and sensitivity for detecting Desulfovibrio DNA in aqueous buffer and biological media. The implementation of this technology for the detection of Desulfovibrio as a tool for monitoring water supply networks is innovative and allows real-time in situ monitoring, making it possible to detect the growth of Desulfovibrio inside of pipes at an early stage and perform timely interventions to reverse it.
Keywords: microbiologically influenced corrosion; molecular gates; nanomaterials; oligonucleotide probe; sulphate-reducing bacteria.
Conflict of interest statement
Authors Román Ponz-Carcelén and María Pedro-Monzonís are employed by the company Global Omnium Group. The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures



References
-
- Kumar M.A., Kasirajan K.T., Parthiban K. Production and characterization of exopolysaccharides (EPS) from biofilm forming marine bacterium. Braz. Arch. Biol. Technol. 2011;54:259–265. doi: 10.1590/S1516-89132011000200006. - DOI
-
- Marschall C., Frenzel P., Cypionka H. Influence of oxygen on sulfate reduction and growth of sulfate-reducing bacteria. Arch. Microbiol. 1993;159:168–173. doi: 10.1007/BF00250278. - DOI
-
- Eguía-López E., Trueba-Ruiz A., Río-Calonge B., Girón-Portilla M.A., Bielva-Tejera C. Recent studies on antifouling systems to artificial structures in marine ecosystem. J. Marit. Res. 2006;3:73–89.
Grants and funding
LinkOut - more resources
Full Text Sources

