Network insights into childhood obesity: unveiling methylated-differentially expressed genes and pathways through integrative bioinformatics analysis
- PMID: 40423250
- PMCID: PMC12147440
- DOI: 10.1530/EC-25-0049
Network insights into childhood obesity: unveiling methylated-differentially expressed genes and pathways through integrative bioinformatics analysis
Abstract
Background: Childhood obesity, a global epidemic with profound impacts on physical and psychological health, remains a complex challenge with elusive underlying mechanisms. This study aimed to unravel the epigenetic landscape of this disease by identifying methylated-differentially expressed genes (MeDEGs) in childhood obesity through integrated bioinformatics approaches.
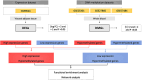
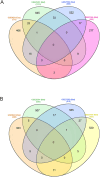
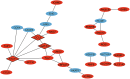
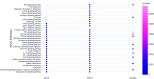
Methods: Expression profiling (GSE9624) and methylation profiling (GSE25301, GSE27860, and GSE57484) datasets containing data on children with obesity (cases) and eutrophic children (control group) were obtained from the Gene Expression Omnibus (GEO) repository. Differentially expressed genes (DEGs) and differentially methylated genes (DMGs) between the groups were identified using GEO2R. MeDEGs were identified by superimposing the lists of DEGs and DMGs. The protein-protein interaction (PPI) network was constructed using the STRING database and analyzed using Cytoscape. Topological and modular PPI network analyses were carried out using the CytoHubba and MCODE plugins, respectively. Functional enrichment analyses were performed based on Gene Ontology terms and KEGG pathways.
Results: A total of 70 MeDEGs were identified, including 45 hypomethylated high-expression and 25 hypermethylated low-expression genes. The PPI network highlighted three hub-bottleneck genes (CCL5, STAT1, and GATA3) and two functional modules. Overall, the 70 MeDEGs were associated with KEGG pathways related to cellular differentiation, inflammation, chemokine signaling, lipid and glucose metabolism, insulin resistance, and apoptosis.
Conclusion: This study, employing integrative bioinformatics approaches, provides insights into the methylation-mediated mechanisms contributing to childhood obesity, advancing our understanding of this multifaceted chronic disease.
Keywords: bioinformatics; childhood obesity; differentially methylated genes; methylation-regulated differentially expressed genes (MeDEGs); systems biology.
Conflict of interest statement
The authors declare that there is no conflict of interest that could be perceived as prejudicing the impartiality of the work reported.
Figures

Similar articles
-
Integrated bioinformatics approach reveals methylation-regulated differentially expressed genes in obesity.Arch Endocrinol Metab. 2023 May 12;67(4):e000604. doi: 10.20945/2359-3997000000604. Arch Endocrinol Metab. 2023. PMID: 37252693 Free PMC article.
-
Comprehensive analysis of DNA methylation and gene expression profiles in cholangiocarcinoma.Cancer Cell Int. 2019 Dec 26;19:352. doi: 10.1186/s12935-019-1080-y. eCollection 2019. Cancer Cell Int. 2019. PMID: 31889904 Free PMC article.
-
Identification of DNA methylation-regulated differentially expressed genes in RA by integrated analysis of DNA methylation and RNA-Seq data.J Transl Med. 2022 Oct 22;20(1):481. doi: 10.1186/s12967-022-03664-5. J Transl Med. 2022. PMID: 36273177 Free PMC article.
-
Identification of aberrantly methylated differentially expressed genes and pro-tumorigenic role of KIF2C in melanoma.Front Genet. 2022 Jul 22;13:817656. doi: 10.3389/fgene.2022.817656. eCollection 2022. Front Genet. 2022. PMID: 35991567 Free PMC article.
-
Exploring nasopharyngeal carcinoma genetics: Bioinformatics insights into pathways and gene associations.Med J Malaysia. 2024 Sep;79(5):615-625. Med J Malaysia. 2024. PMID: 39352166 Review.
References
-
- World Health Organization . Obesity and overweight. Geneva, Switzerland: WHO. (https://www.who.int/news-room/fact-sheets/detail/obesity-and-overweight)
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

