Label-free single-cell phenotyping to determine tumor cell heterogeneity in pancreatic cancer in real time
- PMID: 40424076
- PMCID: PMC12288891
- DOI: 10.1172/jci.insight.169105
Label-free single-cell phenotyping to determine tumor cell heterogeneity in pancreatic cancer in real time
Abstract
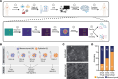
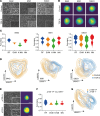
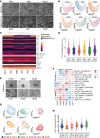
Resistance to chemotherapy of pancreatic ductal adenocarcinoma (PDAC) is largely driven by intratumoral heterogeneity (ITH) due to tumor cell plasticity and clonal diversity. To develop alternative strategies to overcome this defined mechanism of resistance, tools to monitor and quantify ITH in a rapid and scalable fashion are needed urgently. Here, we employed label-free digital holographic microscopy (DHM) to characterize ITH in PDAC. We established a robust experimental and machine learning analysis pipeline to perform single-cell phenotyping based on DHM-derived phase images of PDAC cells in suspension. Importantly, we were able to detect dynamic changes in tumor cell differentiation and heterogeneity of distinct PDAC subtypes upon induction of epithelial-mesenchymal transition and under treatment-imposed pressure in murine and patient-derived model systems. This platform allowed us to assess phenotypic ITH in PDAC on a single-cell level in real time. Implementing this technology into the clinical workflow has the potential to fundamentally increase our understanding of tumor heterogeneity during evolution and treatment response.
Keywords: Cancer; Gastroenterology; Oncology.
Conflict of interest statement
Figures

References
MeSH terms
LinkOut - more resources
Full Text Sources
Medical

