The IL-18 receptor is expressed on murine small-intestinal enterochromaffin cells and executes a recovery program upon injury
- PMID: 40424129
- PMCID: PMC12146721
- DOI: 10.1073/pnas.2417149122
The IL-18 receptor is expressed on murine small-intestinal enterochromaffin cells and executes a recovery program upon injury
Abstract
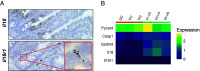
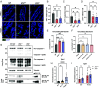
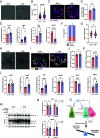
Upon injury, epithelial-derived IL-18 is released and induces an inflammatory response in underlying IL18R1+ lamina propria cells. Notably, Il18r1 is also predicted to be expressed and functional in intestinal epithelial cells (IECs), since epithelial IL18R1 deficiency contributes to worsened outcomes upon inflammatory challenge. However, the nature of Il18r1+ IECs, and their subsequent role in epithelial-intrinsic IL-18 signaling is poorly characterized. Here, we show that, in the murine small intestine, the IL-18 receptor is expressed by rare IECs that we identified to be a subset of enterochromaffin cells (ECC). While these cells are the major producers of serotonin in the intestine, we found no evidence that IL-18 regulated serotonin metabolism or release. Rather, upon radiation-induced injury, Il18r1+ cells appeared in the crypt base and took on a revival stem cell (revSC) program, marked by mixed expression of YAP/TAZ and enteroendocrine genes signatures. Functionally, irradiated Il18-/- mice display reduced epithelial proliferation and altered differentiation in the small intestine, characterized by increased Paneth cells (PC) and elevated Wnt3 levels, which was partially recapitulated in Il18-/- ileal organoids. In sum, we identified an Il18r1+ population in the epithelium and revealed a role for IEC-intrinsic IL-18 signaling during injury.
Keywords: IL-18 receptor; enterochromaffin cells; inflammasomes; intestinal injury; revival stem cells.
Conflict of interest statement
Competing interests statement:The authors declare no competing interest.
Figures



References
-
- Broz P., Dixit V. M., Inflammasomes: Mechanism of assembly, regulation and signalling. Nat. Rev. Immunol. 16, 407–420 (2016). - PubMed
-
- Kayagaki N., et al. , Caspase-11 cleaves gasdermin D for non-canonical inflammasome signalling. Nature 526, 666–671 (2015). - PubMed
-
- Lamkanfi M., Dixit V. M., Mechanisms and functions of inflammasomes. Cell 157, 1013–1022 (2014). - PubMed
-
- Shi J., et al. , Cleavage of GSDMD by inflammatory caspases determines pyroptotic cell death. Nature 526, 660–665 (2015). - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

