YAP controls cell migration and invasion through a Rho GTPase switch
- PMID: 40424361
- PMCID: PMC12268267
- DOI: 10.1126/scisignal.adu3794
YAP controls cell migration and invasion through a Rho GTPase switch
Abstract
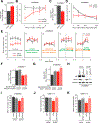
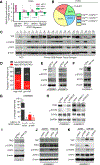
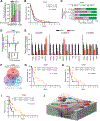
Delineating the mechanisms that control the movement of cells is central to understanding diverse physiological and pathophysiological processes. The transcriptional coactivator YAP is important during development and associated with cancer metastasis. Here, we found that YAP promoted cell migration by modulating a Rho family guanosine triphosphatase (GTPase) switch involving Rac1 and RhoA, which are key regulators of cytoskeletal dynamics. YAP transcriptionally transactivated the gene encoding the Rac1 guanine nucleotide exchange factor TRIO by directly binding to its intronic enhancer. This led to the activation of Rac1 and inhibition of RhoA, which increased cell migration and invasion in vitro and in vivo. This YAP-dependent program was observed across many cell types, including human breast epithelial cells and astrocytes, but it was particularly enhanced in a patient-specific manner in glioblastoma (GBM), the most common malignant brain tumor. Additionally, YAP-TRIO signaling activated STAT3, a transcription factor implicated in invasive growth in cancer, suggesting potential for cross-talk with this pathway to exacerbate invasive behavior. Clinically, hyperactivation of YAP, TRIO, and STAT3 gene signatures in GBM were associated with poor survival outcomes in patients. Our findings suggest that the YAP-TRIO-Rho-GTPase signaling network regulates invasive cell spread in both physiological and pathological contexts.
Conflict of interest statement
Figures
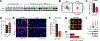



References
-
- Sahai E, Marshall CJ, RHO-GTPases and cancer. Nat Rev Cancer 2, 133–142 (2002). - PubMed
-
- Calvo F, Ege N, Grande-Garcia A, Hooper S, Jenkins RP, Chaudhry SI, Harrington K, Williamson P, Moeendarbary E, Charras G, Sahai E, Mechanotransduction and YAP-dependent matrix remodelling is required for the generation and maintenance of cancer-associated fibroblasts. Nat Cell Biol 15, 637–646 (2013). - PMC - PubMed
-
- Dupont S, Morsut L, Aragona M, Enzo E, Giulitti S, Cordenonsi M, Zanconato F, Le Digabel J, Forcato M, Bicciato S, Elvassore N, Piccolo S, Role of YAP/TAZ in mechanotransduction. Nature 474, 179–183 (2011). - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

