Distinct Regulation of Early Trafficking of the NMDA Receptors by the Ligand-Binding Domains of the GluN1 and GluN2A Subunits
- PMID: 40425318
- PMCID: PMC12199545
- DOI: 10.1523/JNEUROSCI.0226-24.2025
Distinct Regulation of Early Trafficking of the NMDA Receptors by the Ligand-Binding Domains of the GluN1 and GluN2A Subunits
Abstract
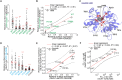
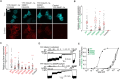
N-Methyl-d-aspartate receptors (NMDARs) play a crucial role in excitatory neurotransmission, with numerous pathogenic variants identified in the GluN subunits, including their ligand-binding domains (LBDs). The prevailing hypothesis postulates that the endoplasmic reticulum (ER) quality control machinery verifies the agonist occupancy of NMDARs, but this was tested in a limited number of studies. Using microscopy and electrophysiology in the human embryonic kidney 293 (HEK293) cells, we found that surface expression of GluN1/GluN2A receptors containing a set of alanine substitutions within the LBDs correlated with the measured EC50 values for glycine (GluN1 subunit mutations) while not correlating with the measured EC50 values for l-glutamate (GluN2A subunit mutations). The mutant cycle of GluN1-S688 residue, including the pathogenic GluN1-S688Y and GluN1-S688P variants, showed a correlation between relative surface expression of the GluN1/GluN2A receptors and the measured EC50 values for glycine, as well as with the calculated ΔG binding values for glycine obtained from molecular dynamics simulations. In contrast, the mutant cycle of GluN2A-S511 residue did not show any correlation between the relative surface expression of the GluN1/GluN2A receptors and the measured EC50 values for l-glutamate or calculated ΔG binding values for l-glutamate. Coexpression of both mutated GluN1 and GluN2A subunits led to additive or synergistic alterations in the surface number of GluN1/GluN2A receptors. The synchronized ER release by ARIAD technology confirmed the altered early trafficking of GluN1/GluN2A receptors containing the mutated LBDs. The microscopical analysis from embryonal rat hippocampal neurons (both sexes) corroborated our conclusions from the HEK293 cells.
Keywords: Golgi apparatus; endoplasmic reticulum; glutamate receptor; hippocampal neuron; ion channel; pathogenic variant.
Copyright © 2025 Netolicky et al.
Conflict of interest statement
The authors declare no competing financial interests.
Figures








References
-
- Berendsen HJC, Postma JPM, van Gunsteren WF, DiNola A, Haak JR (1984) Molecular dynamics with coupling to an external bath. J Chem Phys 81:3684–3690.. 10.1063/1.448118 - DOI
-
- Case DA, et al. (2024) Amber 2024, University of California, San Francisco.
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
