Multiplex base editing to protect from CD33 directed drugs for immune and gene therapy
- PMID: 40425554
- PMCID: PMC12116803
- DOI: 10.1038/s41467-025-59713-2
Multiplex base editing to protect from CD33 directed drugs for immune and gene therapy
Abstract
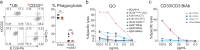
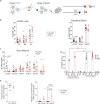
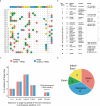
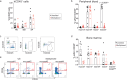
The selection of genetically engineered immune or hematopoietic cells in vivo after gene editing remains a clinical problem and requires a method to spare on-target toxicity to normal cells. Here, we develop a base editing approach exploiting a naturally occurring CD33 single nucleotide polymorphism leading to removal of full-length CD33 surface expression on edited cells. CD33 editing in human and nonhuman primate hematopoietic stem and progenitor cells protects myeloid progeny from CD33-targeted therapeutics without affecting normal hematopoiesis in vivo, thus demonstrating potential for improved immunotherapies with reduced off-leukemia toxicity. For broader application to gene therapies, we demonstrate highly efficient (>70%) multiplexed adenine base editing of the CD33 and gamma globin genes, resulting in long-term persistence of dual gene-edited cells with HbF reactivation in nonhuman primates. Using the CD33 antibody-drug conjugate Gemtuzumab Ozogamicin, we show resistance of engrafted, multiplex edited human cells in vivo, and a 2-fold enrichment for edited cells in vitro. Together, our results highlight the potential of adenine base editors for improved immune and gene therapies.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: This study was funded by a grant from Vor Biopharma and 1R21CA256461 at Columbia University and by grants from NIH/NHLBI (R01 HL136135_2) and NIH/NCI (R01 CA266556) at Fred Hutchinson Cancer Center. Columbia University has licensed technology that is the subject of this study to Vor Biopharma. F.B., A.M.A., and S.M. are coinventors on issued and pending patent applications licensed to Vor Biopharma. S.M. has equity ownership and is on the Scientific Advisory Board of Vor Biopharma. R.B.W. received laboratory research grants and/or clinical trial support from Aptevo, Celgene/Bristol Myers Squibb, ImmunoGen/AbbVie, Janssen, Jazz, Kite, Kura, Pfizer, and Vor Biopharma, and has been a consultant to Wugen. HPK is or was a consultant to and has or had ownership interests with Rocket Pharmaceuticals, Homology Medicines, Vor Biopharma and Ensoma Inc. H-P.K. has also been a consultant to CSL Behring and Magenta Therapeutics. D.R.L. is a consultant for Prime Medicine, Beam Therapeutics, Pairwise Plants, and Chroma Medicine, and Nvelop Therapeutics, companies that use or deliver genome editing or genome engineering agents and owns equity in these companies. The remaining authors declare no competing interests.
Figures




Update of
-
Multiplex Base Editing to Protect from CD33-Directed Therapy: Implications for Immune and Gene Therapy.bioRxiv [Preprint]. 2023 Feb 23:2023.02.23.529353. doi: 10.1101/2023.02.23.529353. bioRxiv. 2023. Update in: Nat Commun. 2025 May 27;16(1):4899. doi: 10.1038/s41467-025-59713-2. PMID: 36865281 Free PMC article. Updated. Preprint.
References
MeSH terms
Substances
Grants and funding
- R01 HL156647/HL/NHLBI NIH HHS/United States
- K99 HL163805/HL/NHLBI NIH HHS/United States
- P51 OD010425/OD/NIH HHS/United States
- R00 HL163805/HL/NHLBI NIH HHS/United States
- R50 CA274319/CA/NCI NIH HHS/United States
- R35 GM118062/GM/NIGMS NIH HHS/United States
- U01 AI142756/AI/NIAID NIH HHS/United States
- R01 CA266556/CA/NCI NIH HHS/United States
- P01 HL053749/HL/NHLBI NIH HHS/United States
- R01 HL136135/HL/NHLBI NIH HHS/United States
- RM1 HG009490/HG/NHGRI NIH HHS/United States
- R01 HL151765/HL/NHLBI NIH HHS/United States
- U42 OD011123/OD/NIH HHS/United States
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

