Chondrocyte fatty acid oxidation drives osteoarthritis via SOX9 degradation and epigenetic regulation
- PMID: 40425566
- PMCID: PMC12117060
- DOI: 10.1038/s41467-025-60037-4
Chondrocyte fatty acid oxidation drives osteoarthritis via SOX9 degradation and epigenetic regulation
Abstract
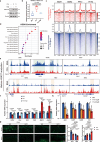
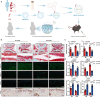
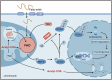
Osteoarthritis is the most prevalent age-related degenerative joint disease and is closely linked to obesity. However, the underlying mechanisms remain unclear. Here we show that altered lipid metabolism in chondrocytes, particularly enhanced fatty acid oxidation (FAO), contributes to osteoarthritis progression. Excessive FAO causes acetyl-CoA accumulation, thereby altering protein-acetylation profiles, where the core FAO enzyme HADHA is hyperacetylated and activated, reciprocally boosting FAO activity and exacerbating OA progression. Mechanistically, elevated FAO reduces AMPK activity, impairs SOX9 phosphorylation, and ultimately promotes its ubiquitination-mediated degradation. Additionally, acetyl-CoA orchestrates epigenetic modulation, affecting multiple cellular processes critical for osteoarthritis pathogenesis, including the transcriptional activation of MMP13 and ADAMTS7. Cartilage-targeted delivery of trimetazidine, an FAO inhibitor and AMPK activator, demonstrates superior efficacy in a mouse model of metabolism-associated post-traumatic osteoarthritis. These findings suggest that targeting chondrocyte-lipid metabolism may offer new therapeutic strategies for osteoarthritis.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures




References
-
- Martel-Pelletier, J. et al. Osteoarthritis. Nat. Rev. Dis. Prim.2, 16072 (2016). - PubMed
-
- Han, S. Osteoarthritis year in review 2022: biology. Osteoarthr. Cartil.30, 1575–1582 (2022). - PubMed
-
- Ng, L. J. et al. SOX9 binds DNA, activates transcription, and coexpresses with type II collagen during chondrogenesis in the mouse. Dev. Biol.183, 108–121 (1997). - PubMed
MeSH terms
Substances
Grants and funding
- LR22H060001/Natural Science Foundation of Zhejiang Province (Zhejiang Provincial Natural Science Foundation)
- Q23H060019/Natural Science Foundation of Zhejiang Province (Zhejiang Provincial Natural Science Foundation)
- 82422044/National Natural Science Foundation of China (National Science Foundation of China)
- 82272522/National Natural Science Foundation of China (National Science Foundation of China)
- U22A20282/National Natural Science Foundation of China (National Science Foundation of China)
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

