Paxilline derived from an endophytic fungus of Baphicacanthus cusia alleviates hepatocellular carcinoma through autophagy-mediated apoptosis
- PMID: 40425675
- PMCID: PMC12117046
- DOI: 10.1038/s41598-025-03044-1
Paxilline derived from an endophytic fungus of Baphicacanthus cusia alleviates hepatocellular carcinoma through autophagy-mediated apoptosis
Abstract
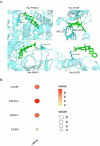
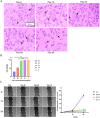
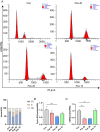
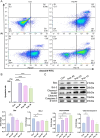
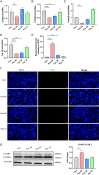
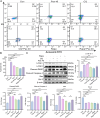
Hepatocellular carcinoma (HCC) is a malignancy for which no effective drugs are available. Paxilline is derived from an endophytic fungus of the leaves of Baphicacanthus cusia (Nees) Bremek. In a previous study, we found that paxilline inhibited the proliferation of HepG2 cells; however, its mechanism remains unclear. In this study, ESI+ and NMR were used to characterize the paxilline structure. Network pharmacology analysis was performed with databases and software to obtain the core targets and signaling pathways associated with the anti-HCC effects of paxilline. Molecular docking was performed to validate the preliminary affinity of paxilline for the core targets. For further in vitro experiments, a CCK8 assay was performed to detect cell viability, a wound healing assay was performed to detect cell migration, an Annexin V-FITC assay was performed to detect the cell cycle and apoptosis rate in HepG2 cells, RT-qPCR analysis was performed to detect the expression of cell cycle-related genes and autophagy-related genes, Immunofluorescence staining was performed to detect the expression of LC3B, and Western blotting was performed to detect the expression of apoptosis-related proteins and autophagy-related proteins. As a result, we obtained a white powder, which was identified as paxilline. Network analysis and molecular docking results revealed that apoptosis-related and autophagy-relatted protein were key targets (mTOR and PI3K) for paxilline anti-HCC. Further examination revealed that paxilline promoted HepG2 cell apoptosis, inhibited HepG2 cell migration, and arrested HepG2 cell in the S phage. RT-qPCR analysis revealed that paxilline markedly downregulated the mRNA expression of Cyclin D1, CDK4, LC3B, mTOR, Parkin, and PINK1. Immunofluorescence staining demonstrated a significant upregulation of LC3B protein expression following paxilline treatment. Further validation by Western blotting showed that paxilline significantly increased the expression of LC3B II/I, bax, cleaved-PARP, and cleaved-caspase 3, while significantly decreased the expression of bcl-2. Additionally, a significant promotion of cellular apoptosis and expression of apoptotic proteins when treatment with chloroquine (CQ)/rapamycin (Rapa). Meanwhile, when combined with paxilline, it was found that paxilline may have a synergistic effects with Rapa, jointly promoting cellular apoptosis and the expression of proapoptotic proteins. In conclusion, these findings reveled paxilline alleviates HCC through autophagy-mediated apoptosis.
Keywords: Apoptosis; Autophagy; Hepatocellular carcinoma; Network pharmacology; Paxilline.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Competing interests: The authors declare no competing interests.
Figures


Similar articles
-
Antitumor effect of matrine in human hepatoma G2 cells by inducing apoptosis and autophagy.World J Gastroenterol. 2010 Sep 14;16(34):4281-90. doi: 10.3748/wjg.v16.i34.4281. World J Gastroenterol. 2010. PMID: 20818811 Free PMC article.
-
Hydrogen sulfide promotes autophagy of hepatocellular carcinoma cells through the PI3K/Akt/mTOR signaling pathway.Cell Death Dis. 2017 Mar 23;8(3):e2688. doi: 10.1038/cddis.2017.18. Cell Death Dis. 2017. PMID: 28333142 Free PMC article.
-
A novel all-trans retinoic acid derivative 4-amino‑2‑trifluoromethyl-phenyl retinate inhibits the proliferation of human hepatocellular carcinoma HepG2 cells by inducing G0/G1 cell cycle arrest and apoptosis via upregulation of p53 and ASPP1 and downregulation of iASPP.Oncol Rep. 2016 Jul;36(1):333-41. doi: 10.3892/or.2016.4795. Epub 2016 May 9. Oncol Rep. 2016. PMID: 27177208
-
JS-K, a nitric oxide prodrug, induces DNA damage and apoptosis in HBV-positive hepatocellular carcinoma HepG2.2.15 cell.Biomed Pharmacother. 2017 Aug;92:989-997. doi: 10.1016/j.biopha.2017.05.141. Epub 2017 Jun 8. Biomed Pharmacother. 2017. PMID: 28605880
-
[Effect of danusertib on cell cycle, apoptosis and autophagy of hepatocellular carcinoma HepG2 cells in vitro].Nan Fang Yi Ke Da Xue Xue Bao. 2018 Dec 30;38(12):1476-1484. doi: 10.12122/j.issn.1673-4254.2018.12.13. Nan Fang Yi Ke Da Xue Xue Bao. 2018. PMID: 30613017 Free PMC article. Chinese.
References
-
- Forner, A., Reig, M. & Bruix, J. Hepatocellular carcinoma. Lancet391, 1301–1314 (2018). - PubMed
-
- Bruix, J., Boix, L., Sala, M. & Llovet, J. M. Focus on hepatocellular carcinoma. Cancer Cell5, 215–219 (2004). - PubMed
-
- Vogel, A., Meyer, T., Sapisochin, G., Salem, R. & Saborowski, A. Hepatocellular carcinoma. Lancet400, 1345–1362 (2022). - PubMed
-
- Yang, C. et al. Evolving therapeutic landscape of advanced hepatocellular carcinoma. Nat. Rev. Gastroenterol. Hepatol.20, 203–222 (2023). - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

