Proteomic surveys of the mouse heart unveil cardiovascular responses to nitric oxide/cGMP signaling deficiencies
- PMID: 40425728
- PMCID: PMC12116794
- DOI: 10.1038/s42003-025-08203-8
Proteomic surveys of the mouse heart unveil cardiovascular responses to nitric oxide/cGMP signaling deficiencies
Abstract
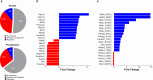
Diminished bioavailability of nitric oxide (NO) contributes to the pathogenesis of cardiometabolic disorders. However, the alterations in signaling under NO deficiency remain mostly unknown. We combined genetics and proteomics to quantify changes in the heart proteome, phosphoproteome, and S-nitrosocysteine proteome in mice lacking nitric oxide synthases (NOS1, NOS2, NOS3), lacking all three enzymes (tNOS), or the alpha 1-regulatory subunit of the soluble guanylate cyclase (sGCα1). Modest changes of less than 1% in the proteome and 4% in the phosphoproteome in single NOS gene or sGCα1 null mouse hearts indicate sufficient biological compensation. In contrast, the number of S-nitrosylated proteins declined by 80%, 57%, and 35% in NOS3, NOS1, and NOS2 null mice, respectively. A 21% remodeling of the proteome and 9% of the phosphoproteome in the tNOS null mice included integral kinases that provide adaptive rewiring of signaling. The data revealed the emergence of enhanced mitogen-activated-kinases Mapk3/Mapk1 signaling, documented by increased phosphorylation of these kinases and their downstream targets. The data highlight that adaptive compensation of signaling prevents overt phenotypes during NO signaling deficits. In contrast, maladaptive signaling via Mapk3/Mapk1 may promote pathological cardiomyopathy that progressively develops in the tNOS null mice.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures





References
-
- Furchgott, R. F. The 1996 Albert Lasker Medical Research Awards. The discovery of endothelium-derived relaxing factor and its importance in the identification of nitric oxide. JAMA276, 1186–1188 (1996). - PubMed
-
- Murad, F. Shattuck Lecture. Nitric oxide and cyclic GMP in cell signaling and drug development. N. Engl. J. Med.355, 2003–2011 (2006). - PubMed
-
- Ignarro, L. J. Nitric oxide: a unique endogenous signaling molecule in vascular biology. Biosci. Rep.19, 51–71 (1999). - PubMed
-
- Lundberg, J. O., Gladwin, M. T. & Weitzberg, E. Strategies to increase nitric oxide signaling in cardiovascular disease. Nat. Rev. Drug Discov.4, 623–641 (2015). - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

