MOTS-c mimics exercise to combat diabetic liver fibrosis by targeting Keap1-Nrf2-Smad2/3
- PMID: 40425777
- PMCID: PMC12116857
- DOI: 10.1038/s41598-025-03526-2
MOTS-c mimics exercise to combat diabetic liver fibrosis by targeting Keap1-Nrf2-Smad2/3
Abstract
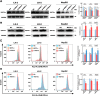
Liver fibrosis is a common complication of T2DM(Type 2 diabetes mellitus). Appropriate intervention (exercise or drugs) in the early stage of liver fibrosis can slow down or even reverse liver fibrosis. MOTS-c (Mitochondrial open reading frame of the 12 S r RNA type-c ) has been described as an exercise-mimicking substance, and its effects are similar to those achieved by aerobic exercise; however, the exact mechanism remains to be elucidated. In this study, liver function was impaired in a T2DM rat model, leading to the aggravation of liver fibrosis. T2DM rats with liver fibrosis were subjected to MOTS-c, aerobic exercise therapy, or their combination. HE staining, Masson's trichrome staining and immunohistochemistry were used for histopathological examination. Transcriptome sequencing, q-PCR and WB were used to detect the expression of Keap1 (Kelch-like ECH-associated protein 1), Nrf2 (Nuclear factor erythroid 2-related factor 2 ), Smad2/3/4 and other genes. MOTS-c and aerobic exercise therapy improved T2DM-induced liver fibrosis. Additionally, cells were transfected with MOTS-c overexpression or interference plasmids or MOTS-c was added to the culture medium. MOTS-c overexpression or MOTS-c addition to the culture medium inhibited ROS levels, increased the mRNA and protein expression of Keap1-Nrf2 pathway genes and decreased the expression of TGF-β1(Transforming growth factor-beta1)/Smad pathway genes. Our findings demonstrate that MOTS-c modulates the progression of T2DM complicated by liver fibrosis through a Keap1-Nrf2-Smad2/3 signaling pathway-dependent mechanism.
Keywords: Aerobic exercise; Keap1-Nrf2-Smad2/3 pathway; Liver fibrosis; MOTS-c; T2DM.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Competing interests: The authors declare no competing interests. Supplementary Information: All of the data is contained within the article and the supplementary materials.
Figures





References
-
- Zheng, Y., Ley, S. H. & Hu, F. B. Global aetiology and epidemiology of type 2 diabetes mellitus and its complications. Nat. Rev. Endocrinol.14, 88–98. 10.1038/nrendo.2017.151 (2018). - PubMed
-
- Younossi, Z. M. et al. The global epidemiology of NAFLD and NASH in patients with type 2 diabetes: A systematic review and meta-analysis. J. Hepatol.71, 793–801. 10.1016/j.jhep.2019.06.021 (2019). - PubMed
MeSH terms
Substances
Grants and funding
- 2024NSFSC1228/Natural Science Foundation of Sichuan Province
- 2023M740381/China Postdoctoral Science Foundation
- GZB20240083/Postdoctoral Fellowship Program (Grade B) of China Postdoctoral Science Foundation
- 2024-A024/Key Laboratory of Sports Medicine of Sichuan Province
- ZYGH2402/Chengdu Sport University 2024-2025 'Excellent Scientific Research Program'
LinkOut - more resources
Full Text Sources
Medical
Research Materials

