Feasibility of multiomics tumor profiling for guiding treatment of melanoma
- PMID: 40425842
- PMCID: PMC12283375
- DOI: 10.1038/s41591-025-03715-6
Feasibility of multiomics tumor profiling for guiding treatment of melanoma
Erratum in
-
Publisher Correction: Feasibility of multiomics tumor profiling for guiding treatment of melanoma.Nat Med. 2025 Aug;31(8):2817-2818. doi: 10.1038/s41591-025-03904-3. Nat Med. 2025. PMID: 40750934 Free PMC article. No abstract available.
Abstract
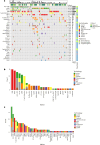
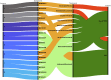
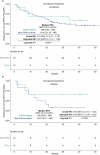
There is limited evidence supporting the feasibility of using omics and functional technologies to inform treatment decisions. Here we present results from a cohort of 116 melanoma patients in the prospective, multicentric observational Tumor Profiler (TuPro) precision oncology project. Nine independent technologies, mostly at single-cell level, were used to analyze 126 patient samples, generating up to 500 Gb of data per sample (40,000 potential markers) within 4 weeks. Among established and experimental markers, the molecular tumor board selected 54 to inform its treatment recommendations. In 75% of cases, TuPro-based data were judged to be useful in informing recommendations. Patients received either standard of care (SOC) treatments or highly individualized, polybiomarker-driven treatments (beyond SOC). The objective response rate in difficult-to-treat palliative, beyond SOC patients (n = 37) was 38%, with a disease control rate of 54%. Progression-free survival of patients with TuPro-informed therapy decisions was 6.04 months, (95% confidence interval, 3.75-12.06) and 5.35 months (95% confidence interval, 2.89-12.06) in ≥third therapy lines. The proof-of-concept TuPro project demonstrated the feasibility and relevance of omics-based tumor profiling to support data-guided clinical decision-making. ClinicalTrials.gov identifier: NCT06463509 .
© 2025. The Author(s).
Conflict of interest statement
Competing interests: B.B. is a founder and shareholder of Navignostics—a precision oncology spin-off from the University of Zurich—based on multiplexed imaging. V.H.K. reports being an invited speaker for Sharing Progress in Cancer Care (SPCC) and Indica Labs; advisory board of Takeda; sponsored research agreements with Roche and IAG, all unrelated to the current study. V.H.K. is a participant in patent applications on the assessment of cancer immunotherapy biomarkers by digital pathology and for the prediction of cancer recurrence risk and prediction of treatment efficacy using deep learning unrelated to the current study. G.G. is listed as inventor on patents related to the 4i technology (WO 2019/207004; WO 2020/008071, EP3922973B1). R.D. received funding from Novartis, Merck Sharp & Dohme (MSD), Bristol-Myers Squibb (BMS), Roche, Amgen, Takeda, Pierre Fabre, Sun Pharma, Sanofi, Catalym, Second Genome, Regeneron, T3 Pharma, MaxiVAX SA, Pfizer and Simcere. M.W. has received research grants from Novartis, Quercis and Versameb, and honoraria for lectures or advisory board participation or consulting from Anheart, Bayer, Curevac, Medac, Neurosense, Novartis, Novocure, Orbus, Pfizer, Philogen, Roche and Servier. A.W. and R.D. were part of the MTB that discussed the TuPro results. The other authors declare no competing interests.
Figures







References
-
- Michielin, O., Van Akkooi, A. C. J., Ascierto, P. A., Dummer, R. & Keilholz, U. Cutaneous melanoma: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann. Oncol.30, 1884–1901 (2019). - PubMed
-
- Coit, D. G. et al. Cutaneous melanoma, version 2.2019, NCCN clinical practice guidelines in oncology. J. Natl Compr. Cancer Netw.17, 367–402 (2019). - PubMed
-
- Rocque, G. B. et al. Concordance with NCCN treatment guidelines: relations with health care utilization, cost, and mortality in breast cancer patients with secondary metastasis. Cancer124, 4231–4240 (2018). - PubMed
-
- Andreano, A., Rebora, P., Valsecchi, M. G. & Russo, A. G. Adherence to guidelines and breast cancer patients survival: a population-based cohort study analyzed with a causal inference approach. Breast Cancer Res Treat.164, 119–131 (2017). - PubMed
Publication types
MeSH terms
Substances
Associated data
LinkOut - more resources
Full Text Sources
Medical

