A tritrophic plant-insect-pathogen system used to develop a closely linked Rag2 and Rsv1-h recombinant haplotype in double-resistant soybean germplasm
- PMID: 40426039
- PMCID: PMC12117769
- DOI: 10.1186/s12864-025-11686-8
A tritrophic plant-insect-pathogen system used to develop a closely linked Rag2 and Rsv1-h recombinant haplotype in double-resistant soybean germplasm
Abstract
Background: The colocalization of two resistance (R) genes on chromosome 13 of soybean (Glycine max (L.) Merrill) that confer resistance against the soybean aphid (Aphis glycines) and soybean mosaic virus (SMV) gives rise to a very unique R-avr tritrophic incompatible interaction system that goes across biological kingdoms. In this tritrophic system, the insect is the only natural vector of the virus and soybean is a host-plant for both pests/pathogen. The almost unavoidable co-evolution of pathogen-vector with that of the R-genes in soybean plants through an endless arms race to avoid each other's defense-attack mechanisms raises interesting questions. The objectives of this work were to (i) develop double-resistant recombinant inbred lines (RILs) with a Rag2-Rsv1-h gene haplotype in coupling phase using resistance alleles from two different genetic sources (PI 243540 (Rag2) and Suweon 97 (Rsv1-h)), (ii) confirm phenotypically the resistant reaction against both pests in double-resistant RILs, and (iii) dissect the Rag2-Rsv1-h region with molecular markers and investigate the potential for structural variation.
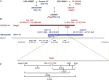
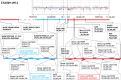
Results: We observed a recombination event in identified double-resistant F3:5 RILs in a region of chromosome 13 ca. 21 kb long (between positions 30,297,227 and 30,318,949 in Wm82.a2.v1) that lies between the reported locations of the Rsv1-h and Rag2 genes (29,815,463--29,912,369 and 30,412,581--30,466,533 intervals, respectively, based on Wm82.a2.v1), indicating the double-resistant haplotype is in coupling phase. The tight LD estimates obtained between haplotype markers underscored the physical proximity of the two resistance genes. Only 10 recombinant haplotype classes (excluding double heterozygotes) were observed among the 51 that were possible with a four loci haplotype. The 10 recombinant classes represented 15 out of 192 screened individuals. A joint SMV-aphid phenotypic greenhouse screen allowed us to identify the best aphid biotype 1 and SMV-G1, double resistant haplotype class in recombinant progeny. Our molecular marker results agree with previous fine-mapping reports and preclude the presence of resistance genes other than Rag2 and Rsv1-h in double-resistant RILs. A comparative genomic hybridization analysis revealed no obvious structural variants in the region.
Conclusions: To our knowledge, this is the first report of double-resistant Rag2-Rsv1-h soybean RILs that used a plant-insect-pathogen tritrophic system for germplasm enhancement. The co-occurrence of Rag and Rsv genes in a region that clusters resistance genes on chromosome 13 may be a unique feature of domesticated soybean. The recombinant genotypes will be useful in breeding to develop soybean cultivars with resistance to both the vector and the virus. The parental and recombinant genotypes may be helpful in future studies to elucidate interesting evolutionary questions regarding vector, host, and virus tritrophic systems.
Keywords: R-gene; Breeding; Candidate genes; Comparative genomic hybridization; Crop improvement; Linkage; Recombinant haplotype; Resistance; Single nucleotide polymorphism; Soybean (Glycine max (L.) Merrill).
© 2025. The Author(s).
Conflict of interest statement
Declarations. Ethics approval and consent to participate: Not applicable. Consent for publication: Not applicable. Competing interests: The authors declare no competing interests.
Figures

Similar articles
-
Fine mapping of the Rsv1-h gene in the soybean cultivar Suweon 97 that confers resistance to two Chinese strains of the soybean mosaic virus.Theor Appl Genet. 2016 Nov;129(11):2227-2236. doi: 10.1007/s00122-016-2769-0. Epub 2016 Aug 20. Theor Appl Genet. 2016. PMID: 27544525
-
Multiplex single nucleotide polymorphism (SNP) assay for detection of soybean mosaic virus resistance genes in soybean.Theor Appl Genet. 2011 Feb;122(2):445-57. doi: 10.1007/s00122-010-1459-6. Epub 2010 Oct 8. Theor Appl Genet. 2011. PMID: 20931170
-
Genetic mapping of three quantitative trait loci for soybean aphid resistance in PI 567324.Heredity (Edinb). 2013 Jul;111(1):16-22. doi: 10.1038/hdy.2013.10. Epub 2013 Mar 13. Heredity (Edinb). 2013. PMID: 23486080 Free PMC article.
-
A bean common mosaic virus (BCMV)-resistance gene is fine-mapped to the same region as Rsv1-h in the soybean cultivar Suweon 97.Theor Appl Genet. 2018 Sep;131(9):1851-1860. doi: 10.1007/s00122-018-3117-3. Epub 2018 Jun 16. Theor Appl Genet. 2018. PMID: 29909526
-
The Resistant Soybean-Aphis glycines Interaction: Current Knowledge and Prospects.Front Plant Sci. 2020 Aug 11;11:1223. doi: 10.3389/fpls.2020.01223. eCollection 2020. Front Plant Sci. 2020. PMID: 32849757 Free PMC article. Review.
References
-
- Soy Stats. American Soybean Association. https://soygrowers.com/education-resources/publications/soy-stats/. Accessed 1 October 2024.
-
- USDA-ERS. National Agricultural Statistics Service Homepage. https://www.nass.usda.gov/index.php. Accessed 10 September 2024.
-
- USDA ERS - Oil Crops Sector at a Glance. https://www.ers.usda.gov/topics/crops/soybeans-and-oil-crops/oil-crops-s.... Accessed 10 September 2024.
-
- FAO. Food and Agriculture Organization of the United Nations. 2011. Country report on the state of plant genetic resources for food and agriculture. United States of America. http://www.fao.org/fileadmin/templates/agphome/documents/PGR/SoW2/countr.... Accessed 10 September 2024.
MeSH terms
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

