The genetic structure and diversity of smallholder dairy cattle in Rwanda
- PMID: 40426054
- PMCID: PMC12107919
- DOI: 10.1186/s12863-025-01323-4
The genetic structure and diversity of smallholder dairy cattle in Rwanda
Abstract
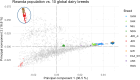
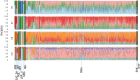
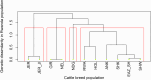
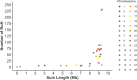
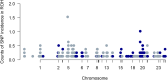
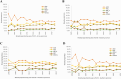
Previous genomic characterisation of Rwanda dairy cattle predominantly focused on the One Cow per Poor Family (locally called "Girinka") programme. However, smallholder farmers in Rwanda have benefited from other livestock initiatives and development programmes. Capturing and documenting the genetic diversity, is critical in part as a key contribution to genomic resource required to support dairy development in Rwanda. A total of 2,229 crossbred animals located in all dairy-producing regions of Rwanda were sampled. For each animal, a hair sample was collected and genotyped by using the Geneseek Genomic Profiler (GGP, Neogen Geneseek®) Bovine 50 K (n = 1,917) and GGP Bovine 100 K arrays (n = 312). The combined dataset was subject to quality control, data curation for use in population genetics and genomic analyses. To assess the genetic structure and diversity of the current population, key analyses for population structure were applied: Principal Component Analysis (PCA), population structure and diversity, admixture analysis, measures of heterozygosity, runs of homozygosity (ROH) and minor allelic frequency (MAF). A dataset of global dairy population of European taurine, African indicus and African taurus (n = 250) was used as reference. Results showed that Rwanda cattle population is highly admixed of diverse pure and crossbred animals with average MAF of 33% (standard error; se = 0.001) with proportion of foreign high yielding (taurine) dairy breeds of Jersey Island (18%); 12% non-Island Jersey and 42% Holstein-Friesian ancestries. Two African Bos taurus and five Bos indicus breeds contributed 28% of their genetics. Genetic distances were highest in Gir and N'dama (0.29); and Nelore and N'dama (0.29). There were 1,331 ROH regions and average heterozygosity were high for Rwanda cattle (0.41 se = 0.001). Asides well-established genes in cattle, we found evidence for a variety of novel and less-known genes under selection to be associated with fertility, milk production, innate immunity and environmental adaptation. This observed diversity offers opportunity to decipher the presence and/or lack of genetic variations to initiate short- and long-term breed improvement programmes for adaptation traits, disease resistance, heat tolerance, productivity and profitability of smallholder dairy systems in Rwanda.
Keywords: Admixture; Runs of homozygosity; Rwanda population structure; SNP arrays; Smallholder dairy.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Ethics approval and consent to participate: This study was approved by the ethics committee of the University of Rwanda’s Research and Postgraduate Studies (RPGS) unit in accordance with the 1964 Helsinki declaration and its later amendments or comparable ethical standards. Consent for publication: Not applicable. Competing interests: The authors declare no competing interests.
Figures



Similar articles
-
Genomic diversity of Cameroonian Gudali and Gudali-cross cattle.Sci Rep. 2025 Apr 29;15(1):15066. doi: 10.1038/s41598-025-99799-8. Sci Rep. 2025. PMID: 40301666 Free PMC article.
-
Use of High Density Single Nucleotide Polymorphism (SNP) Arrays to Assess Genetic Diversity and Population Structure of Dairy Cattle in Smallholder Dairy Systems: The Case of Girinka Programme in Rwanda.Front Genet. 2018 Oct 10;9:438. doi: 10.3389/fgene.2018.00438. eCollection 2018. Front Genet. 2018. PMID: 30364216 Free PMC article.
-
The patterns of admixture, divergence, and ancestry of African cattle populations determined from genome-wide SNP data.BMC Genomics. 2020 Dec 7;21(1):869. doi: 10.1186/s12864-020-07270-x. BMC Genomics. 2020. PMID: 33287702 Free PMC article.
-
Immunogenetic influences on tick resistance in African cattle with particular reference to trypanotolerant N'Dama (Bos taurus) and trypanosusceptible Gobra zebu (Bos indicus) cattle.Acta Trop. 2000 May 31;75(3):263-77. doi: 10.1016/s0001-706x(00)00063-2. Acta Trop. 2000. PMID: 10838210 Review.
-
Heat stress effects on milk yield traits and metabolites and mitigation strategies for dairy cattle breeds reared in tropical and sub-tropical countries.Front Vet Sci. 2023 Jul 7;10:1121499. doi: 10.3389/fvets.2023.1121499. eCollection 2023. Front Vet Sci. 2023. PMID: 37483284 Free PMC article. Review.
References
-
- Ben Jemaa S, Rahal O, Gaouar SBS, Mastrangelo S, Boussaha M, Ciani E. Genomic characterization of Algerian guelmoise cattle and their genetic relationship with other North African populations inferred from SNP genotyping arrays. Livest Sci. 2018;217:19–25. 10.1016/j.livsci.2018.09.009.
-
- Wilson R. Crossbreeding of cattle in Africa. J Agric Environ Sci. 2018;6. 10.15640/JAES.V7N1A3.
-
- Rahal O, Aissaoui C, Ata N, Yilmaz O, Cemal I, Ameur Ameur A, Gaouar SBS. Genetic characterization of four Algerian cattle breeds using microsatellite markers. Anim Biotechnol. 2020. 10.1080/10495398.2020.1746321. - PubMed
-
- Mehdid B, Benyarou M, Ameur Ameur A, Gaouar SBS. Contribution to the study of two local bovine breeds in Wilaya of Tlemcen: morphometric characterisation and DNA biobank. Genet Biodivers J. 2018;2:44–55. 10.46325/gabj.v2i1.114.
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
