DNA binding effects of LDH nanozyme for aseptic osteolysis mitigation through STING pathway modulation
- PMID: 40426255
- PMCID: PMC12117809
- DOI: 10.1186/s12951-025-03458-z
DNA binding effects of LDH nanozyme for aseptic osteolysis mitigation through STING pathway modulation
Abstract
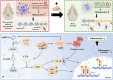
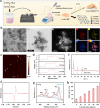
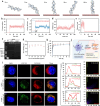
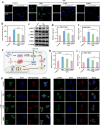
Persistent and intense inflammation is recognized as the primary cause of wear-particle-induced aseptic osteolysis, which ultimately resulting in aseptic prosthesis loosening. Reducing inflammation plays a significant role in mitigating osteolysis, and the STING pathway has emerged as a promising therapeutic target for its prevention. Specifically, damaged periprosthetic cells of aseptic osteolysis release double-stranded DNA (dsDNA) into the osteolytic microenvironment, serving as a specific stimulus for the STING pathway. Herein, we found that layered double hydroxide (LDH) nanozyme exhibited a robust DNA-binding capacity primarily mediated by van der Waals interactions, which showed superior performance in inhibiting dsDNA-induced inflammation of aseptic osteolysis. Importantly, such binding capability enabled effective co-loading LDH with STING inhibitor C176, thus facilitating inhibition of the STING pathway. Such synergistic actions contributed to ameliorate the inflammatory milieu and remodel the osteolysis microenvironment successfully to reduce cranial bone damage, which was confirmed on animal model of osteolysis. Collectively, this strategy demonstrated an effective approach by utilizing synergistic effects to establish a positive feedback loop in the treatment of osteolysis, thereby alleviating TiPs-induced periprosthetic osteolysis and preventing postoperative complications.
Keywords: Inflammation; Nanozyme; Osteolysis; Oxidative stress; cGAS-STING pathway.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Ethics and consent to publish: All animal experiments were authorized by the Ethics Committee of the Shanghai Jiao Tong University (A2023128-001). Competing interests: The authors declare no competing interests.
Figures


Similar articles
-
Osthole ameliorates wear particle-induced osteogenic impairment by mitigating endoplasmic reticulum stress via PERK signaling cascade.Mol Med. 2024 Dec 20;30(1):266. doi: 10.1186/s10020-024-01034-z. Mol Med. 2024. PMID: 39707212 Free PMC article.
-
Ultrasound-activated erythrocyte membrane-camouflaged Pt (II) layered double hydroxide enhances PD-1 inhibitor efficacy in triple-negative breast cancer through cGAS-STING pathway-mediated immunogenic cell death.Theranostics. 2025 Jan 2;15(4):1456-1477. doi: 10.7150/thno.102284. eCollection 2025. Theranostics. 2025. PMID: 39816689 Free PMC article.
-
STING/TBK1 Regulates Inflammation in Macrophages and Titanium Particles-Induced Osteolysis.ACS Biomater Sci Eng. 2023 Jun 12;9(6):3273-3284. doi: 10.1021/acsbiomaterials.2c01509. Epub 2023 May 3. ACS Biomater Sci Eng. 2023. PMID: 37134278
-
Mechanism of regulating macrophages/osteoclasts in attenuating wear particle-induced aseptic osteolysis.Front Immunol. 2023 Oct 4;14:1274679. doi: 10.3389/fimmu.2023.1274679. eCollection 2023. Front Immunol. 2023. PMID: 37860014 Free PMC article. Review.
-
STING-Pathway Inhibiting Nanoparticles (SPINs) as a Platform for Treatment of Inflammatory Diseases.ACS Appl Bio Mater. 2024 Aug 19;7(8):4867-4878. doi: 10.1021/acsabm.3c01305. Epub 2024 Apr 2. ACS Appl Bio Mater. 2024. PMID: 38563162 Free PMC article. Review.
References
-
- Ormsby RT, Solomon LB, Yang D, Crotti TN, Haynes DR, Findlay DM, Atkins GJ. Osteocytes respond to particles of clinically-relevant conventional and cross-linked polyethylene and metal alloys by up-regulation of resorptive and inflammatory pathways. Acta Biomater. 2019;87:296–306. - PubMed
-
- Springer BD, Sotile WM. The psychology of total joint arthroplasty. J GROMACS Arthroplasty. 2020;35(6S):S46–9. - PubMed
-
- Tian Y, Terkawi MA, Onodera T, Alhasan H, Matsumae G, Takahashi D, Hamasaki M, Ebata T, Aly MK, Kida H, Shimizu T, Uetsuki K, Kadoya K, Iwasaki N. Blockade of XCL1/Lymphotactin ameliorates severity of periprosthetic osteolysis triggered by Polyethylene-Particles. Front Immunol. 2020;11:1720. - PMC - PubMed
-
- Fritz EA, Glant TT, Vermes C, Jacobs JJ, Roebuck KA. Chemokine gene activation in human bone marrow-derived osteoblasts following exposure to particulate wear debris. J Biomed Mater Res A. 2006;77(1):192–201. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

