Characteristics of ESBL-positive Klebsiella pneumoniae isolated from paired children with and without diarrhea
- PMID: 40426260
- PMCID: PMC12117907
- DOI: 10.1186/s13099-025-00700-9
Characteristics of ESBL-positive Klebsiella pneumoniae isolated from paired children with and without diarrhea
Abstract
Background: Klebsiella pneumoniae producing extended-spectrum β-lactamase (ESBL) colonizing and transmitting in the intestine, especially in children, have significant public health implications. Investigating antibiotic resistance, antibiotic resistance genes (ARGs), virulence factor genes (VFGs), and genetic relationships may help us to explore the characteristics and differences of ESBL-positive K. pneumoniae in children with and without diarrhea.
Methods: After selecting and pairing, 26 pairs of 52 ESBL-positive K. pneumoniae strains were isolated from 323 children with diarrhea and 393 children without diarrhea. Antimicrobial susceptibility test and whole genome sequencing were performed to explore antibiotic resistance, ARGs, and VFGs. The genetic relationship was explored by conducting a maximum likelihood phylogenetic tree and investigating plasmid and sequence type (ST).
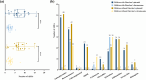
Results: All strains showed resistance to cephalosporins, with ESBL-producing genes widely carried (98.1%). Carbapenem-resistant K. pneumoniae (CRKP) were found in both groups. Hypervirulent K. pneumoniae (hvKP) were isolated from children with diarrhea carrying iucA on plasmid. The emergence of ST5670 CRKP and ST2108 hvKP highlighted the necessity for close monitoring of community-acquired K. pneumoniae.
Conclusions: Severe drug resistance was found among ESBL-positive K. pneumoniae strains isolated from children with and without diarrhea. Attention must be paid to ESBL-positive K. pneumoniae colonized in the intestine of children, and pathogen and ARG monitoring in children should be strengthened, even in healthy people.
Keywords: Klebsiella pneumoniae; Children; Diarrhea; ESBL.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Ethics approval and consent to participate: The study was conducted in accordance with the Declaration of Helsinki, and approved by Ethical Committee of National Institute for communicable disease control and prevention (protocol code ICDC-2017003, 3 Jul. 2017) (protocol code ICDC-2017004, 3 Jul. 2017). Consent for publication: Not applicable. Competing interests: The authors declare no competing interests. Informed consent: Informed consent was obtained from all subjects involved in the study.
Figures


Similar articles
-
Outer Membrane Vesicles Transmitting blaNDM-1 Mediate the Emergence of Carbapenem-Resistant Hypervirulent Klebsiella pneumoniae.Antimicrob Agents Chemother. 2023 May 17;67(5):e0144422. doi: 10.1128/aac.01444-22. Epub 2023 Apr 13. Antimicrob Agents Chemother. 2023. PMID: 37052502 Free PMC article.
-
Molecular characteristics of extended-spectrum β-lactamase-producing Klebsiella pneumoniae in Japan: Predominance of CTX-M-15 and emergence of hypervirulent clones.Int J Infect Dis. 2020 Sep;98:281-286. doi: 10.1016/j.ijid.2020.06.083. Epub 2020 Jun 30. Int J Infect Dis. 2020. PMID: 32619765
-
Relationship between virulence and carbapenem resistance phenotype of Klebsiella pneumoniae from blood infection: identification of a carbapenem-resistant and hypervirulent strain.Zhejiang Da Xue Xue Bao Yi Xue Ban. 2024 Aug 25;53(4):490-497. doi: 10.3724/zdxbyxb-2024-0104. Zhejiang Da Xue Xue Bao Yi Xue Ban. 2024. PMID: 39183061 Free PMC article. Chinese, English.
-
Mobile genetic elements encoding antibiotic resistance genes and virulence genes in Klebsiella pneumoniae: important pathways for the acquisition of virulence and resistance.Front Microbiol. 2025 Feb 24;16:1529157. doi: 10.3389/fmicb.2025.1529157. eCollection 2025. Front Microbiol. 2025. PMID: 40066273 Free PMC article. Review.
-
Epidemiological characteristics and molecular evolution mechanisms of carbapenem-resistant hypervirulent Klebsiella pneumoniae.Front Microbiol. 2022 Sep 12;13:1003783. doi: 10.3389/fmicb.2022.1003783. eCollection 2022. Front Microbiol. 2022. PMID: 36188002 Free PMC article. Review.
References
-
- World Health Organization. Diarrhoeal disease 2024 [Available from: https://www.who.int/news-room/fact-sheets/detail/diarrhoeal-disease
-
- GBD 2015 Mortality and Causes of Death Collaborators. Global, regional, and National life expectancy, all-cause mortality, and cause-specific mortality for 249 causes of death, 1980–2015: a systematic analysis for the global burden of disease study 2015. Lancet. 2016;388(10053):1459–544. - PMC - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources

