Phenotypic and Genotypic Characterization of ESBL-, AmpC-, and Carbapenemase-Producing Klebsiella pneumoniae and High-Risk Escherichia coli CC131, with the First Report of ST1193 as a Causative Agent of Urinary Tract Infections in Human Patients in Algeria
- PMID: 40426551
- PMCID: PMC12108494
- DOI: 10.3390/antibiotics14050485
Phenotypic and Genotypic Characterization of ESBL-, AmpC-, and Carbapenemase-Producing Klebsiella pneumoniae and High-Risk Escherichia coli CC131, with the First Report of ST1193 as a Causative Agent of Urinary Tract Infections in Human Patients in Algeria
Abstract
Background: High-risk Escherichia coli clones, such as sequence type (ST)131 and ST1193, along with multidrug-resistant (MDR) Klebsiella pneumoniae, are globally recognized for their significant role in urinary tract infections (UTIs). This study aimed to provide an overview of the virulence factors, clonal diversity, and antibiotic resistance profiles of extended-spectrum cephalosporin (ESC)-E. coli and K. pneumoniae causing UTIs in humans in the Tebessa region of Algeria.
Methods: Forty E. coli and 17 K. pneumoniae isolates exhibiting ESC-resistance were recovered (July 2022-January 2024) from urine samples of patients at three healthcare facilities to be phenotypically and genotypically characterized. Whole genome sequencing (WGS) was performed on the ST1193 clone.
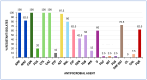
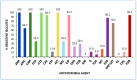
Results: Among K. pneumoniae isolates, all except one harbored CTX-M-15, with a single isolate carrying blaCTX-M-194. Additionally, two K. pneumoniae isolates co-harboring blaCTX-M-15 and blaNDM exhibited phenotypic and genotypic hypervirulence traits. Fluoroquinolone resistance (FQR) was detected in 94.1% of K. pneumoniae isolates. The E. coli isolates carried diverse ESC-resistance genes, including CTX-M-15 (87.5%), CTX-M-27 (5%), CTX-M-1, CMY-59, and CMY-166 (2.5% each). Co-carriage of blaESC and blaOXA-48 was identified in three E. coli isolates, while 62.5% exhibited FQR. Phylogenetic analysis revealed that 52.5% of E. coli belonged to phylogroup B2, including the high-risk clonal complex (CC)131 CH40-30 (17 isolates) and ST1193 (one isolate). In silico analysis of the ST1193 genome determined O75:H5-B2 (CH14-64), and the carriage of IncI1-I(Alpha) and IncF [F-:A1:B10] plasmids. Notably, core genome single-nucleotide polymorphism (SNP) analysis demonstrated high similarity between the Algerian ST1193 isolate and a previously annotated genome from a hospital in Northwest Spain.
Conclusions: This study highlights the spread and genetic diversity of E. coli CC131 CH40-30 and hypervirulent K. pneumoniae clones in Algeria. It represents the first report of a CTX-M-15-carrying E. coli ST1193 in the region. The findings emphasize the urgent need for antibiotic optimization programs and enhanced surveillance to curb the dissemination of high-risk clones that pose an increasing public health threat in Algeria. A simplified method based on virulence traits for E. coli and K. pneumoniae is proposed here for antimicrobial resistance (AMR) monitoring.
Keywords: Algeria; ESBL; Escherichia coli; Klebsiella pneumoniae; ST1193; ST131; carbapenemases; urinary tract infection (UTI).
Conflict of interest statement
The authors declare no conflicts of interest.
Figures

Similar articles
-
Fluoroquinolone resistance in complicated urinary tract infections: association with the increased occurrence and diversity of Escherichia coli of clonal complex 131, together with ST1193.Front Cell Infect Microbiol. 2024 Feb 27;14:1351618. doi: 10.3389/fcimb.2024.1351618. eCollection 2024. Front Cell Infect Microbiol. 2024. PMID: 38510968 Free PMC article.
-
Occurrence and Genomic Characterization of Clone ST1193 Clonotype 14-64 in Uncomplicated Urinary Tract Infections Caused by Escherichia coli in Spain.Microbiol Spectr. 2022 Jun 29;10(3):e0004122. doi: 10.1128/spectrum.00041-22. Epub 2022 May 23. Microbiol Spectr. 2022. PMID: 35604206 Free PMC article.
-
Extended Spectrum β-Lactamase-Producing Escherichia coli and Klebsiella pneumoniae from Broiler Liver in the Center of Algeria, with Detection of CTX-M-55 and B2/ST131-CTX-M-15 in Escherichia coli.Microb Drug Resist. 2021 Feb;27(2):268-276. doi: 10.1089/mdr.2020.0024. Epub 2020 Jun 29. Microb Drug Resist. 2021. PMID: 32609048
-
Mapping Antimicrobial Resistance in Escherichia coli and Klebsiella pneumoniae from Complicated Urinary Tract Infections in Oman: Phenotypic and Genotypic Insights.Diagnostics (Basel). 2025 Apr 22;15(9):1062. doi: 10.3390/diagnostics15091062. Diagnostics (Basel). 2025. PMID: 40361883 Free PMC article.
-
Escherichia coli ST1193: Following in the Footsteps of E. coli ST131.Antimicrob Agents Chemother. 2022 Jul 19;66(7):e0051122. doi: 10.1128/aac.00511-22. Epub 2022 Jun 6. Antimicrob Agents Chemother. 2022. PMID: 35658504 Free PMC article. Review.
References
Grants and funding
LinkOut - more resources
Full Text Sources

